Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline
- PMID: 18053747
- PMCID: PMC2861339
- DOI: 10.1016/j.neuroimage.2007.10.031
Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline
Abstract
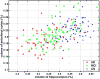
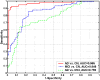
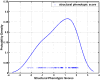
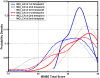
Spatial patterns of brain atrophy in mild cognitive impairment (MCI) and Alzheimer's disease (AD) were measured via methods of computational neuroanatomy. These patterns were spatially complex and involved many brain regions. In addition to the hippocampus and the medial temporal lobe gray matter, a number of other regions displayed significant atrophy, including orbitofrontal and medial-prefrontal grey matter, cingulate (mainly posterior), insula, uncus, and temporal lobe white matter. Approximately 2/3 of the MCI group presented patterns of atrophy that overlapped with AD, whereas the remaining 1/3 overlapped with cognitively normal individuals, thereby indicating that some, but not all, MCI patients have significant and extensive brain atrophy in this cohort of MCI patients. Importantly, the group with AD-like patterns presented much higher rate of MMSE decline in follow-up visits; conversely, pattern classification provided relatively high classification accuracy (87%) of the individuals that presented relatively higher MMSE decline within a year from baseline. High-dimensional pattern classification, a nonlinear multivariate analysis, provided measures of structural abnormality that can potentially be useful for individual patient classification, as well as for predicting progression and examining multivariate relationships in group analyses.
Figures







References
-
- Adeli H, Ghosh-Dastidar S, Dadmehr N. Alzheimer’s disease and models of computation: imaging, classification, and neural models. Journal of Alzheimer’s Disease. 2005;7:187–199. - PubMed
-
- Ashburner J, Csernansky JG, Davatzikos C, Fox NC, Frisoni GB, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. The Lancet (Neurology) 2003;2:79–88. - PubMed
-
- Bennett SAL, Pappas BA, Stevens WD, Davidson CM, Fortin T, Chen J. Cleavage of amyloid precursor protein elicited by chronic cerebral hypoperfusion. Neurobiology of Aging. 2000;21:207–214. - PubMed
-
- Beresford T, Arciniegas D, Alfers J, Clapp L, Martin B, Beresford H, Du Y, Liu D, Shen D, Davatzikos C, Laudenslager M. Hypercortisolism In Alcohol Dependence and Its Relation to Hippocampal Volume Loss. Journal of Studies on Alcohol. 2006;67:861–867. - PubMed
-
- Beresford TP, Arciniegas DB, Alfers J, Clapp L, Martin B, Du Y, Liu D, Shen D, Davatzikos C. Hippocampus Volume Loss Due to Chronic Heavy Drinking. Alcoholism: Clinical and Experimental Research. 2006;30:1866–1870. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

