Persistent transcription of a nonintegrating mutant of simian immunodeficiency virus in rhesus macrophages
- PMID: 18054062
- PMCID: PMC2350229
- DOI: 10.1016/j.virol.2007.11.002
Persistent transcription of a nonintegrating mutant of simian immunodeficiency virus in rhesus macrophages
Abstract
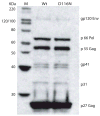
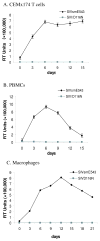
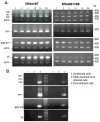
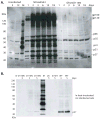
A nonintegrating mutant, SIVsmD116N, was derived from the infectious pathogenic SIVsmE543-3 clone by introducing an Asp (D) to Asn (N) mutation into the catalytic domain of integrase. Although SIVsmD116N generated all viral proteins following transfection, cell-free virus did not productively infect CEMx174 cells, macaque peripheral blood mononuclear cells (PBMCs) or monocyte-derived macrophages (MDM). Viral DNA and transcripts were observed transiently in SIVsmD116N-infected CEMx174 cells and macaque PBMC but persisted in MDM for as long as 20 days. Circular forms of viral DNA were detected but there was no evidence of integration detected by Alu PCR. We found that SIV D116N mutant remained transcriptionally active and expressed low levels of viral proteins persistently in MDM. These data are consistent with a role for macrophages as a persistent latent reservoir for AIDS viruses. The capacity of nonintegrating SIV to persistently generate viral products in macrophages suggests that nonintegrating lentiviral vectors could be engineered to efficiently and safely express proteins for vaccine purposes.
Figures


Similar articles
-
Reduced Simian Immunodeficiency Virus Replication in Macrophages of Sooty Mangabeys Is Associated with Increased Expression of Host Restriction Factors.J Virol. 2015 Oct;89(20):10136-44. doi: 10.1128/JVI.00710-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202248 Free PMC article.
-
Antibodies that neutralize SIV(mac)251 in T lymphocytes cause interruption of the viral life cycle in macrophages by preventing nuclear import of viral DNA.Virology. 2001 Sep 1;287(2):436-45. doi: 10.1006/viro.2001.1053. Virology. 2001. PMID: 11531420
-
Use of helper-free replication-defective simian immunodeficiency virus-based vectors to study macrophage and T tropism: evidence for distinct levels of restriction in primary macrophages and a T-cell line.J Virol. 2001 Mar;75(5):2288-300. doi: 10.1128/JVI.75.5.2288-2300.2001. J Virol. 2001. PMID: 11160732 Free PMC article.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
-
Brain Macrophages in Simian Immunodeficiency Virus-Infected, Antiretroviral-Suppressed Macaques: a Functional Latent Reservoir.mBio. 2017 Aug 15;8(4):e01186-17. doi: 10.1128/mBio.01186-17. mBio. 2017. PMID: 28811349 Free PMC article.
Cited by
-
Nonintegrating Lentiviral Vector-Based Vaccine Efficiently Induces Functional and Persistent CD8+ T Cell Responses in Mice.J Biomed Biotechnol. 2010;2010:534501. doi: 10.1155/2010/534501. Epub 2010 May 19. J Biomed Biotechnol. 2010. PMID: 20508727 Free PMC article.
-
Generation of macrophages from peripheral blood monocytes in the rhesus monkey.J Immunol Methods. 2009 Dec 31;351(1-2):36-40. doi: 10.1016/j.jim.2009.09.013. Epub 2009 Oct 8. J Immunol Methods. 2009. PMID: 19818793 Free PMC article.
-
Immunologic and Virologic Mechanisms for Partial Protection from Intravenous Challenge by an Integration-Defective SIV Vaccine.Viruses. 2017 Jun 2;9(6):135. doi: 10.3390/v9060135. Viruses. 2017. PMID: 28574482 Free PMC article.
-
Development and use of SIV-based Integrase defective lentiviral vector for immunization.Vaccine. 2009 Jul 23;27(34):4622-9. doi: 10.1016/j.vaccine.2009.05.070. Epub 2009 Jun 11. Vaccine. 2009. PMID: 19523909 Free PMC article.
-
An HIV-1 replication pathway utilizing reverse transcription products that fail to integrate.J Virol. 2013 Dec;87(23):12701-20. doi: 10.1128/JVI.01939-13. Epub 2013 Sep 18. J Virol. 2013. PMID: 24049167 Free PMC article.
References
-
- Cara A, Vargas J, Jr, Keller M, Jones S, Mosoian A, Gurtman A, Cohen A, Parkas V, Wallach F, Chusid E, Gelman IH, Klotman ME. Circular viral DNA and anomalous junction sequence in PBMC of HIV-infected individuals with no detectable plasma HIV RNA. Virology. 2002;292(1):1–5. - PubMed
-
- Chun TW, Davey RT, Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, Fauci AS. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. 2000;6(7):757–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

