Electron detachment dissociation of dermatan sulfate oligosaccharides
- PMID: 18055211
- PMCID: PMC2696562
- DOI: 10.1016/j.jasms.2007.10.007
Electron detachment dissociation of dermatan sulfate oligosaccharides
Abstract
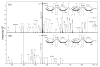
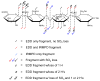
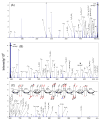
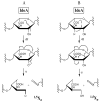
The structural characterization of glycosaminoglycans (GAG) oligosaccharides has been a long-standing challenge in the field of mass spectrometry. In this work, we present the application of electron detachment dissociation (EDD) Fourier transform mass spectrometry to the analysis of dermatan sulfate (DS) oligosaccharides up to 10 residues long. The EDD mass spectra of DS oligosaccharides were compared with their infrared multiphoton dissociation (IRMPD) mass spectra. EDD produces more abundant fragmentation than IRMPD with far less loss of SO3 from labile sulfate modifications. EDD cleaves all glycosidic bonds, yielding both conventional glycosidic bond fragmentation as well as satellite peaks resulting from the additional loss of 1 or 2 hydrogen atoms. EDD also yields more cross-ring fragmentation than IRMPD. For EDD, abundant cross-ring fragmentation in the form of A- and X-ions is observed, with 1,5Xn cleavages occurring for all IdoA residues and many of the GalNAc4S residues, except at the reducing and nonreducing ends. In contrast, IRMPD produces only A-type cross-ring fragmentation for long oligosaccharides (dp6-dp10). As all the structurally informative fragment ions observed by IRMPD appear as a subset of the peaks found in the EDD mass spectrum, EDD shows great potential for the characterization of GAG oligosaccharides using a single tandem mass spectrometry experiment.
Figures




Similar articles
-
Electron detachment dissociation of neutral and sialylated oligosaccharides.J Am Soc Mass Spectrom. 2007 Dec;18(12):2162-72. doi: 10.1016/j.jasms.2007.09.007. Epub 2007 Sep 14. J Am Soc Mass Spectrom. 2007. PMID: 17962039
-
Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides.J Am Soc Mass Spectrom. 2008 Jun;19(6):790-8. doi: 10.1016/j.jasms.2008.03.010. Epub 2008 Apr 3. J Am Soc Mass Spectrom. 2008. PMID: 18499037 Free PMC article.
-
Electron-induced dissociation of glycosaminoglycan tetrasaccharides.J Am Soc Mass Spectrom. 2008 Oct;19(10):1449-58. doi: 10.1016/j.jasms.2008.06.024. Epub 2008 Jul 2. J Am Soc Mass Spectrom. 2008. PMID: 18657442 Free PMC article.
-
Modern developments in mass spectrometry of chondroitin and dermatan sulfate glycosaminoglycans.Amino Acids. 2011 Jul;41(2):235-56. doi: 10.1007/s00726-010-0682-4. Epub 2010 Jul 15. Amino Acids. 2011. PMID: 20632047 Review.
-
Infrared multiphoton dissociation mass spectrometry for structural elucidation of oligosaccharides.Methods Mol Biol. 2009;534:23-35. doi: 10.1007/978-1-59745-022-5_2. Methods Mol Biol. 2009. PMID: 19277545 Review.
Cited by
-
Negative Electron Transfer Dissociation Sequencing of 3-O-Sulfation-Containing Heparan Sulfate Oligosaccharides.J Am Soc Mass Spectrom. 2018 Jun;29(6):1262-1272. doi: 10.1007/s13361-018-1907-0. Epub 2018 Mar 21. J Am Soc Mass Spectrom. 2018. PMID: 29564812 Free PMC article.
-
De Novo Sequencing of Complex Mixtures of Heparan Sulfate Oligosaccharides.Anal Chem. 2016 May 17;88(10):5299-307. doi: 10.1021/acs.analchem.6b00519. Epub 2016 Apr 27. Anal Chem. 2016. PMID: 27087275 Free PMC article.
-
Hexuronic acid stereochemistry determination in chondroitin sulfate glycosaminoglycan oligosaccharides by electron detachment dissociation.J Am Soc Mass Spectrom. 2012 Sep;23(9):1488-97. doi: 10.1007/s13361-012-0428-5. Epub 2012 Jul 24. J Am Soc Mass Spectrom. 2012. PMID: 22825742 Free PMC article.
-
Electron detachment dissociation of fluorescently labeled sialylated oligosaccharides.Electrophoresis. 2011 Dec;32(24):3526-35. doi: 10.1002/elps.201100327. Epub 2011 Nov 24. Electrophoresis. 2011. PMID: 22120881 Free PMC article.
-
Differential Characterization and Classification of Tissue Specific Glycosaminoglycans by Tandem Mass Spectrometry and Statistical Methods.Int J Mass Spectrom. 2012 Feb 15;312:144-154. doi: 10.1016/j.ijms.2011.07.019. Epub 2011 Jul 23. Int J Mass Spectrom. 2012. PMID: 22523474 Free PMC article.
References
-
- Perrimon N, Bernfield M. Cellular functions of proteoglycans--an overview. Semin Cell Dev Biol. 2001;12:65–67. - PubMed
-
- Fannon M, Forsten KE, Nugent MA. Potentiation and Inhibition of bFGF Binding by Heparin: A Model for Regulation of Cellular Response. Biochemistry. 2000;39:1434–1445. - PubMed
-
- Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The Involvement of Heparan Sulfate (HS) in FGF1/HS/FGFR1 Signaling Complex. J Biol Chem. 2003;278:17121–17129. - PubMed
-
- Sadir R, Imberty A, Baleux F, Lortat-Jacob H. Heparan Sulfate/Heparin Oligosaccharides Protect Stromal Cell-derived Factor-1 (SDF-1)/CXCL12 against Proteolysis Induced by CD26/Dipeptidyl Peptidase IV. J Biol Chem. 2004;279:43854–43860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

