microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells
- PMID: 18055230
- PMCID: PMC4135426
- DOI: 10.1016/j.immuni.2007.10.009
microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells
Abstract
microRNA-155 (miR-155) is expressed by cells of the immune system after activation and has been shown to be required for antibody production after vaccination with attenuated Salmonella. Here we show the intrinsic requirement for miR-155 in B cell responses to thymus-dependent and -independent antigens. B cells lacking miR-155 generated reduced extrafollicular and germinal center responses and failed to produce high-affinity IgG1 antibodies. Gene-expression profiling of activated B cells indicated that miR-155 regulates an array of genes with diverse function, many of which are predicted targets of miR-155. The transcription factor Pu.1 is validated as a direct target of miR155-mediated inhibition. When Pu.1 is overexpressed in wild-type B cells, fewer IgG1 cells are produced, indicating that loss of Pu.1 regulation is a contributing factor to the miR-155-deficient phenotype. Our results implicate post-transcriptional regulation of gene expression for establishing the terminal differentiation program of B cells.
Figures
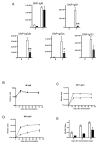
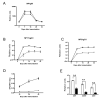
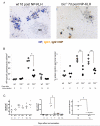

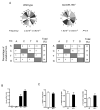
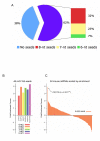
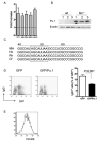
Comment in
-
MicroRNA-155 function in B Cells.Immunity. 2007 Dec;27(6):825-7. doi: 10.1016/j.immuni.2007.11.010. Immunity. 2007. PMID: 18093533
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Cattoretti G, Shaknovich R, Smith PM, Jack HM, Murty VV, Alobeid B. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–6939. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G117/424/MRC_/Medical Research Council/United Kingdom
- MC_U105178806/MRC_/Medical Research Council/United Kingdom
- G0700287/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/00001206/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 079643/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/0000M206/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G8402371/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/0000C223/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000M940/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

