Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1
- PMID: 18055603
- PMCID: PMC2174879
- DOI: 10.1105/tpc.107.054890
Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1
Abstract
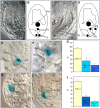
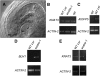
In Arabidopsis thaliana, the female gametophyte is a highly polarized structure consisting of four cell types: one egg cell and two synergids, one central cell, and three antipodal cells. In this report, we describe the characterization of a novel female gametophyte mutant, eostre, which affects establishment of cell fates in the mature embryo sac. The eostre phenotype is caused by misexpression of the homeodomain gene BEL1-like homeodomain 1 (BLH1) in the embryo sac. It is known that BELL-KNAT proteins function as heterodimers whose activities are regulated by the Arabidopsis ovate family proteins (OFPs). We show that the phenotypic effect of BLH1 overexpression is dependent upon the class II knox gene KNAT3, suggesting that KNAT3 must be expressed and functional during megagametogenesis. Moreover, disruption of At OFP5, a known interactor of KNAT3 and BLH1, partially phenocopies the eostre mutation. Our study indicates that suppression of ectopic activity of BELL-KNOX TALE complexes, which might be mediated by At OFP5, is essential for normal development and cell specification in the Arabidopsis embryo sac. As eostre-1 embryo sacs also show nuclear migration abnormalities, this study suggests that a positional mechanism might be directing establishment of cell fates in early megagametophyte development.
Figures




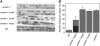
References
-
- Adler, R. (2000). A model of retinal cell differentiation in the chick embryo. Prog. Retin. Eye Res. 19 529–557. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Betschinger, J., and Knoblich, J.A. (2004). Dare to be different: Asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14 R674–R685. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

