Network inference, analysis, and modeling in systems biology
- PMID: 18055607
- PMCID: PMC2174897
- DOI: 10.1105/tpc.107.054700
Network inference, analysis, and modeling in systems biology
Figures

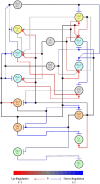
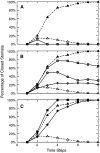
References
-
- Agrawal, V., Zhang, C., Shapiro, A.D., and Dhurjati, P.S. (2004). A dynamic mathematical model to clarify signaling circuitry underlying programmed cell death control in Arabidopsis disease resistance. Biotechnol. Prog. 20 426–442. - PubMed
-
- Albert, I., and Albert, R. (2004). Conserved network motifs allow protein-protein interaction prediction. Bioinformatics 20 3346–3352. - PubMed
-
- Albert, R., and Barabási, A.L. (2002). Statistical mechanics of complex networks. Rev. Mod. Phys. 74 47–97.
-
- Albert, R., DasGupta, B., Dondi, R., Kachalo, S., Sontag, E., Zelikovsky, A., and Westbrooks, K. (2007. a). A novel method for signal transduction network inference from indirect experimental evidence. J. Comput. Biol. 14 927–949. - PubMed
-
- Albert, R., DasGupta, B., Dondi, R., Kachalo, S., Sontag, E.D., Zelikovsky, A., and Westbrook, K. (2007. b). A novel method for signal transduction network inference from indirect experimental evidence. J. Comput. Biol. 14 927–949. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

