Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function
- PMID: 18056081
- PMCID: PMC2241850
- DOI: 10.1093/nar/gkm1057
Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function
Abstract
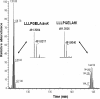
Post-translational modification of histones and other chromosomal proteins regulates chromatin conformation and gene activity. Methylation and acetylation of lysyl residues are among the most frequently described modifications in these proteins. Whereas these modifications have been studied in detail, very little is known about a recently discovered chemical modification, the N(epsilon)-lysine formylation, in histones and other nuclear proteins. Here we mapped, for the first time, the sites of lysine formylation in histones and several other nuclear proteins. We found that core and linker histones are formylated at multiple lysyl residues located both in the tails and globular domains of histones. In core histones, formylation was found at lysyl residues known to be involved in organization of nucleosomal particles that are frequently acetylated and methylated. In linker histones and high mobility group proteins, multiple formylation sites were mapped to residues with important role in DNA binding. N(epsilon)-lysine formylation in chromosomal proteins is relatively abundant, suggesting that it may interfere with epigenetic mechanisms governing chromatin function, which could lead to deregulation of the cell and disease.
Figures


References
-
- Chu F, Nusinow DA, Chalkley RJ, Plath K, Panning B, Burlingame AL. Mapping post-translational modifications of the histone variant MacroH2A1 using tandem mass spectrometry. Mol. Cell. Proteomics. 2006;5:194–203. - PubMed
-
- Medzihradszky KF, Zhang X, Chalkley RJ, Guan S, McFarland MA, Chalmers MJ, Marshall AG, Diaz RL, Allis CD, et al. Characterization of Tetrahymena histone H2B variants and posttranslational populations by electron capture dissociation (ECD) Fourier transform ion cyclotron mass spectrometry (FT-ICR MS) Mol. Cell. Proteomics. 2004;3:872–886. - PubMed
-
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. - PubMed
-
- Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, et al. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. - PubMed
-
- Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol. Cell. Proteomics. 2006;5:541–552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

