Multiple, conserved cryptic recombination signals in VH gene segments: detection of cleavage products only in pro B cells
- PMID: 18056287
- PMCID: PMC2150985
- DOI: 10.1084/jem.20071224
Multiple, conserved cryptic recombination signals in VH gene segments: detection of cleavage products only in pro B cells
Abstract
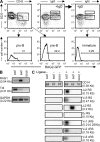
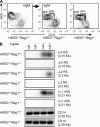
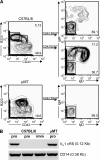
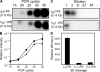
Receptor editing is believed to play the major role in purging newly formed B cell compartments of autoreactivity by the induction of secondary V(D)J rearrangements. In the process of immunoglobulin heavy (H) chain editing, these secondary rearrangements are mediated by direct V(H)-to-J(H) joining or cryptic recombination signals (cRSs) within V(H) gene segments. Using a statistical model of RS, we have identified potential cRSs within V(H) gene segments at conserved sites flanking complementarity-determining regions 1 and 2. These cRSs are active in extrachromosomal recombination assays and cleaved during normal B cell development. Cleavage of multiple V(H) cRSs was observed in the bone marrow of C57BL/6 and RAG2:GFP and microMT congenic animals, and we determined that cRS cleavage efficiencies are 30-50-fold lower than a physiological RS. cRS signal ends are abundant in pro-B cells, including those recovered from microMT mice, but undetectable in pre- or immature B cells. Thus, V(H) cRS cleavage regularly occurs before the generation of functional preBCR and BCR. Conservation of cRSs distal from the 3' end of V(H) gene segments suggests a function for these cryptic signals other than V(H) gene replacement.
Figures
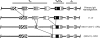
References
-
- Fanning, L., F.E. Bertrand, C. Steinberg, and G.E. Wu. 1998. Molecular mechanisms involved in receptor editing at the Ig heavy chain locus. Int. Immunol. 10:241–246. - PubMed
-
- Chen, C., Z. Nagy, E.L. Prak, and M. Weigert. 1995. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 3:747–755. - PubMed
-
- Kleinfield, R.W., and M.G. Weigert. 1989. Analysis of VH gene replacement events in a B cell lymphoma. J. Immunol. 142:4475–4482. - PubMed
-
- Sakano, H., Y. Kurosawa, M. Weigert, and S. Tonegawa. 1981. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 290:562–565. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

