Rap1a null mice have altered myeloid cell functions suggesting distinct roles for the closely related Rap1a and 1b proteins
- PMID: 18056377
- PMCID: PMC2722108
- DOI: 10.4049/jimmunol.179.12.8322
Rap1a null mice have altered myeloid cell functions suggesting distinct roles for the closely related Rap1a and 1b proteins
Erratum in
- J Immunol. 2008 Mar 1;180(5):3612. Munugulavadla, Veerendra [corrected to Munugalavadla, Veerendra]
Abstract
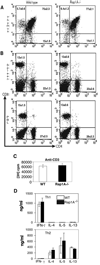
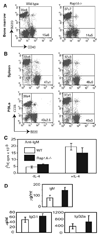
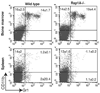
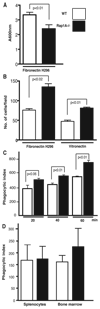
The Ras-related GTPases Rap1a and 1b have been implicated in multiple biological events including cell adhesion, free radical production, and cancer. To gain a better understanding of Rap1 function in mammalian physiology, we deleted the Rap1a gene. Although loss of Rap1a expression did not initially affect mouse size or viability, upon backcross into C57BL/6J mice some Rap1a-/- embryos died in utero. T cell, B cell, or myeloid cell development was not disrupted in Rap1a-/- mice. However, macrophages from Rap1a null mice exhibited increased haptotaxis on fibronectin and vitronectin matrices that correlated with decreased adhesion. Chemotaxis of lymphoid and myeloid cells in response to CXCL12 or CCL21 was significantly reduced. In contrast, an increase in FcR-mediated phagocytosis was observed. Because Rap1a was previously copurified with the human neutrophil NADPH oxidase, we addressed whether GTPase loss affected superoxide production. Neutrophils from Rap1a-/- mice had reduced fMLP-stimulated superoxide production as well as a weaker initial response to phorbol ester. These results suggest that, despite 95% amino acid sequence identity, similar intracellular distribution, and broad tissue distribution, Rap1a and 1b are not functionally redundant but rather differentially regulate certain cellular events.
Conflict of interest statement
The authors have no financial conflict of interest.
Figures
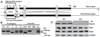

Similar articles
-
Small GTPases Rap1 and RhoA regulate superoxide formation by Rac1 GTPases activation during the phagocytosis of IgG-opsonized zymosans in macrophages.Free Radic Biol Med. 2012 May 1;52(9):1796-805. doi: 10.1016/j.freeradbiomed.2012.02.004. Epub 2012 Feb 11. Free Radic Biol Med. 2012. PMID: 22330068
-
Rap1a is a key regulator of fibroblast growth factor 2-induced angiogenesis and together with Rap1b controls human endothelial cell functions.Mol Cell Biol. 2008 Sep;28(18):5803-10. doi: 10.1128/MCB.00393-08. Epub 2008 Jul 14. Mol Cell Biol. 2008. PMID: 18625726 Free PMC article.
-
Functional redundancy between RAP1 isoforms in murine platelet production and function.Blood. 2018 Nov 1;132(18):1951-1962. doi: 10.1182/blood-2018-03-838714. Epub 2018 Aug 21. Blood. 2018. PMID: 30131434 Free PMC article.
-
Divergent Functions of Rap1A and Rap1B in Endothelial Biology and Disease.Int J Mol Sci. 2025 Jun 4;26(11):5372. doi: 10.3390/ijms26115372. Int J Mol Sci. 2025. PMID: 40508181 Free PMC article. Review.
-
Rap1 GTPases: an emerging role in the cardiovasculature.Life Sci. 2011 Apr 11;88(15-16):645-52. doi: 10.1016/j.lfs.2011.01.023. Epub 2011 Feb 2. Life Sci. 2011. PMID: 21295042 Free PMC article. Review.
Cited by
-
Distinct functions for Rap1 signaling in vascular morphogenesis and dysfunction.Exp Cell Res. 2013 Sep 10;319(15):2350-9. doi: 10.1016/j.yexcr.2013.07.022. Epub 2013 Aug 2. Exp Cell Res. 2013. PMID: 23911990 Free PMC article. Review.
-
Novel role of NADPH oxidase in angiogenesis and stem/progenitor cell function.Antioxid Redox Signal. 2009 Oct;11(10):2517-33. doi: 10.1089/ars.2009.2582. Antioxid Redox Signal. 2009. PMID: 19309262 Free PMC article. Review.
-
Small GTPase Rap1 Is Essential for Mouse Development and Formation of Functional Vasculature.PLoS One. 2015 Dec 29;10(12):e0145689. doi: 10.1371/journal.pone.0145689. eCollection 2015. PLoS One. 2015. PMID: 26714318 Free PMC article.
-
The EPAC-Rap1 pathway prevents and reverses cytokine-induced retinal vascular permeability.J Biol Chem. 2018 Jan 12;293(2):717-730. doi: 10.1074/jbc.M117.815381. Epub 2017 Nov 20. J Biol Chem. 2018. PMID: 29158262 Free PMC article.
-
Isoform-specific differences between Rap1A and Rap1B GTPases in the formation of endothelial cell junctions.Small GTPases. 2011 Mar;2(2):65-76. doi: 10.4161/sgtp.2.2.15735. Small GTPases. 2011. PMID: 21776404 Free PMC article.
References
-
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. - PubMed
-
- Pizon V, Chardin P, Lerosey I, Olofsson B, Tavitian A. Human cDNAs rap1 and rap2 homologous to the Drosophila gene Dras3 encode proteins closely related to ras in the 'effector' region. Oncogene. 1988;3:201–204. - PubMed
-
- Bokoch GM, Parkos CA. Identification of novel GTP-binding proteins in the human neutrophil. FEBS Lett. 1988;227:66–70. - PubMed
-
- Kawata M, Matsui Y, Kondo J, Hishida T, Teranishi Y, Takai Y. A novel small molecular weight GTP-binding protein with the same putative effector domain as the ras proteins in bovine brain membranes. Purification, determination of primary structure, and characterization. J. Biol. Chem. 1988;263:18965–18971. - PubMed
-
- Quinn MT, Parkos CA, Walker L, Orkin SH, Dinauer MC, Jesaitis AJ. Association of a Ras-related protein with cytochrome b of human neutrophils. Nature. 1989;342:198–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

