Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho
- PMID: 18056429
- PMCID: PMC2081983
- DOI: 10.1101/gad.1607807
Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho
Abstract
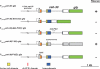
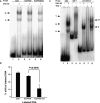
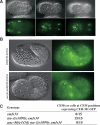
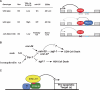
Apoptosis is essential for proper development and tissue homeostasis in metazoans. It plays a critical role in generating sexual dimorphism by eliminating structures that are not needed in a specific sex. The molecular mechanisms that regulate sexually dimorphic apoptosis are poorly understood. Here we report the identification of the ceh-30 gene as a key regulator of sex-specific apoptosis in Caenorhabditis elegans. Loss-of-function mutations in ceh-30 cause the ectopic death of male-specific CEM neurons. ceh-30 encodes a BarH homeodomain protein that acts downstream from the terminal sex determination gene tra-1, but upstream of, or in parallel to, the cell-death-initiating gene egl-1 to protect CEM neurons from undergoing apoptosis in males. The second intron of the ceh-30 gene contains two adjacent cis-elements that are binding sites for TRA-1A and a POU-type homeodomain protein UNC-86 and acts as a sensor to regulate proper specification of the CEM cell fate. Surprisingly, the N terminus of CEH-30 but not its homeodomain is critical for CEH-30's cell death inhibitory activity in CEMs and contains a conserved eh1/FIL domain that is important for the recruitment of the general transcriptional repressor UNC-37/Groucho. Our study suggests that ceh-30 defines a critical checkpoint that integrates the sex determination signal TRA-1 and the cell fate determination and survival signal UNC-86 to control the sex-specific activation of the cell death program in CEMs through the general transcription repressor UNC-37.
Figures


References
-
- Aoki M.P., Aoki A., Maldonado C.A., Aoki A., Maldonado C.A., Maldonado C.A. Sexual dimorphism of apoptosis in lactotrophs induced by bromocryptine. Histochem. Cell Biol. 2001;116:215–222. - PubMed
-
- Argenton F., Ramoz N., Charlet N., Bernardini S., Colombo L., Bortolussi M., Ramoz N., Charlet N., Bernardini S., Colombo L., Bortolussi M., Charlet N., Bernardini S., Colombo L., Bortolussi M., Bernardini S., Colombo L., Bortolussi M., Colombo L., Bortolussi M., Bortolussi M. Mechanisms of transcriptional activation of the promoter of the rainbow trout prolactin gene by GHF1/Pit1 and glucocorticoid. Biochem. Biophys. Res. Commun. 1996;224:57–66. - PubMed
-
- Bae Y.K., Shimizu T., Yabe T., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Shimizu T., Yabe T., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Yabe T., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Nojima H., Muraoka O., Hirano T., Hibi M., Muraoka O., Hirano T., Hibi M., Hirano T., Hibi M., Hibi M. A homeobox gene, pnx, is involved in the formation of posterior neurons in zebrafish. Development. 2003;130:1853–1865. - PubMed
-
- Barr M.M., Sternberg P.W., Sternberg P.W. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. - PubMed
-
- Blush J., Lei J., Ju W., Silbiger S., Pullman J., Neugarten J., Lei J., Ju W., Silbiger S., Pullman J., Neugarten J., Ju W., Silbiger S., Pullman J., Neugarten J., Silbiger S., Pullman J., Neugarten J., Pullman J., Neugarten J., Neugarten J. Estradiol reverses renal injury in Alb/TGF-β1 transgenic mice. Kidney Int. 2004;66:2148–2154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
