CYP2E1 substrate inhibition. Mechanistic interpretation through an effector site for monocyclic compounds
- PMID: 18056994
- PMCID: PMC3933162
- DOI: 10.1074/jbc.M707630200
CYP2E1 substrate inhibition. Mechanistic interpretation through an effector site for monocyclic compounds
Erratum in
- J Biol Chem. 2013 Nov 8;288(45):32640
Abstract
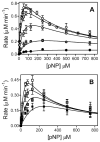
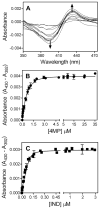
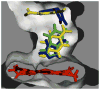
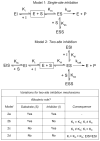
In this study we offer a mechanistic interpretation of the previously known but unexplained substrate inhibition observed for CYP2E1. At low substrate concentrations, p-nitrophenol (pNP) was rapidly turned over (47 min(-1)) with relatively low K(m) (24 microM); nevertheless, at concentrations of >100 microM, the rate of pNP oxidation gradually decreased as a second molecule bound to CYP2E1 through an effector site (K(ss) = 260 microm), which inhibited activity at the catalytic site. 4-Methylpyrazole (4MP) was a potent inhibitor for both sites through a mixed inhibition mechanism. The K(i) for the catalytic site was 2.0 microM. Although we were unable to discriminate whether an EIS or ESI complex formed, the respective inhibition constants were far lower than K(ss). Bicyclic indazole (IND) inhibited catalysis through a single CYP2E1 site (K(i) = 0.12 microM). Similarly, 4MP and IND yielded type II binding spectra that reflected the association of either two 4MP or one IND molecule(s) to CYP2E1, respectively. Based on computational docking studies with a homology model for CYP2E1, the two sites for monocyclic molecules, pNP and 4MP, exist within a narrow channel connecting the active site to the surface of the enzyme. Because of the presence of the heme iron, one site supports catalysis, whereas the other more distal effector site binds molecules that can influence the binding orientation and egress of molecules for the catalytic site. Although IND did not bind these sites simultaneously, the presence of IND at the catalytic site blocked binding at the effector site.
Figures
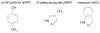

References
-
- Guengerich FP. Toxicol Lett. 1994;70:133–138. - PubMed
-
- Ronis M, Lindros KO, Ingelman-Sundberg M. In: Cytochromes P450, Metabolic and Toxicological Aspects. Ioannides C, editor. CRC Press, Inc; Boca Raton, FL: 1996. pp. 211–239.
-
- Robertson G, Leclercq I, Farrell GC. Am J Physiol. 2001;281:G1135–G1139. - PubMed
-
- Banerjee S, Shang TQ, Wilson AM, Moore AL, Strand SE, Gordon MP, Doty SL. Biotechnol Bioeng. 2002;77:462–466. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

