Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis
- PMID: 18057123
- PMCID: PMC2224249
- DOI: 10.1128/JCM.01468-07
Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis
Abstract
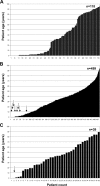
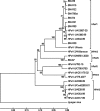
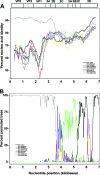
Parechovirus epidemiology and disease association are not fully understood. Real-time reverse transcriptase PCR (RT-PCR) for all human parechoviruses (HPeV) was applied on stool samples from two groups of patients. Both groups contained patients with acute enteritis of all age groups, seen during one full year. Patients with norovirus, adenovirus, enterovirus, astrovirus, or rotavirus infections were excluded. In 118 patients from outbreak and hospital settings, no HPeV was detected. In a prospective study group of 499 nonhospitalized patients, the detection rate was 1.6%. One virus-positive patient was detected from 39 control patients. Positive samples occurred only in summer and autumn. Only one patient had accompanying respiratory symptoms. An association with travel or animal contact was not found. All positive patients except one were <2 years of age, with a neutral gender ratio. In children <2 years of age, the detection rate was 11.6% (7 of 60 children). The range of viral loads was 3,170 to 503,377,290 copies per gram or milliliter of stool. One of the highest viral loads occurred in a control child without symptoms at the time of testing. Phylogenetic analysis showed mainly contemporary HPeV1 strains in our patients but also showed a separate new lineage of HPeV1 in evolutionary transition from the historical prototype strain. Moreover, a novel sixth HPeV type was identified. Full genome analysis of the two viruses revealed recombination between HPeV1 and -3 in one and HPeV6 and -1 in another. HPeV seems relevant in children <2 years and specific RT-PCR for HPeV should be included in enteritis screening.
Figures
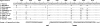
References
-
- Abed, Y., and G. Boivin. 2005. Molecular characterization of a Canadian human parechovirus (HPeV)-3 isolate and its relationship to other HPeVs. J. Med. Virol. 77566-570. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

