Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses
- PMID: 18057136
- PMCID: PMC2238069
- DOI: 10.1128/JCM.01739-07
Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses
Abstract
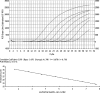
Human rhinoviruses (HRVs) are important contributors to respiratory disease, but their healthcare burden remains unclear, primarily because of the lack of sensitive, accurate, and convenient means of determining their causal role. To address this, we developed and clinically validated the sensitivity and specificity of a real-time reverse transcription-PCR (RT-PCR) assay targeting the viral 5' noncoding region defined by sequences obtained from all 100 currently recognized HRV prototype strains and 85 recently circulating field isolates. The assay successfully amplified all HRVs tested and could reproducibly detect 50 HRV RNA transcript copies, with a dynamic range of over 7 logs. In contrast, a quantified RNA transcript of human enterovirus 68 (HEV68) that showed the greatest sequence homology to the HRV primers and probe set was not detected below a concentration of 5 x 10(5) copies per reaction. Nucleic acid extracts of 111 coded respiratory specimens that were culture positive for HRV or HEV were tested with the HRV real-time RT-PCR assay and by two independent laboratories that used different in-house HRV/HEV RT-PCR assays. Eighty-seven HRV-culture-positive specimens were correctly identified by the real-time RT-PCR assay, and 4 of the 24 HEV-positive samples were positive for HRV. HRV-specific sequences subsequently were identified in these four specimens, suggesting HRV/HEV coinfection in these patients. The assay was successfully applied in an investigation of a coincidental outbreak of HRV respiratory illness among laboratory staff.
Figures


References
-
- Billaud, G., S. Peny, V. Legay, B. Lina, and M. Valette. 2003. Detection of rhinovirus and enterovirus in upper respiratory tract samples using a multiplex nested PCR. J. Virol. Methods 108223-228. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

