Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3
- PMID: 18059169
- PMCID: PMC3164855
- DOI: 10.4161/auto.5278
Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3
Abstract
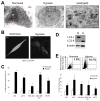
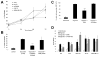
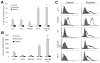
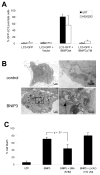
Hypoxia (lack of oxygen) is a physiological stress often associated with solid tumors. Hypoxia correlates with poor prognosis since hypoxic regions within tumors are considered apoptosisresistant. Autophagy (cellular "self digestion") has been associated with hypoxia during cardiac ischemia and metabolic stress as a survival mechanism. However, although autophagy is best characterized as a survival response, it can also function as a mechanism of programmed cell death. Our results show that autophagic cell death is induced by hypoxia in cancer cells with intact apoptotic machinery. We have analyzed two glioma cell lines (U87, U373), two breast cancer cell lines (MDA-MB-231, ZR75) and one embryonic cell line (HEK293) for cell death response in hypoxia (<1% O(2)). Under normoxic conditions, all five cell lines undergo etoposide-induced apoptosis whereas hypoxia fails to induce these apoptotic responses. All five cell lines induce an autophagic response and undergo cell death in hypoxia. Hypoxia-induced cell death was reduced upon treatment with the autophagy inhibitor 3-methyladenine, but not with the caspase inhibitor z-VAD-fmk. By knocking down the autophagy proteins Beclin-1 or ATG5, hypoxia-induced cell death was also reduced. The pro-cell death Bcl-2 family member BNIP3 (Bcl-2/adenovirus E1B 19kDainteracting protein 3) is upregulated during hypoxia and is known to induce autophagy and cell death. We found that BNIP3 overexpression induced autophagy, while expression of BNIP3 siRNA or a dominant-negative form of BNIP3 reduced hypoxia-induced autophagy. Taken together, these results suggest that prolonged hypoxia induces autophagic cell death in apoptosis-competent cells, through a mechanism involving BNIP3.
Conflict of interest statement
I have no conflict of interest or financial disclosure statements to declare.
Figures


References
-
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. - PubMed
-
- Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. - PubMed
-
- Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. - PubMed
-
- Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, Kimchi A. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
