Determinants of RNA quality from FFPE samples
- PMID: 18060057
- PMCID: PMC2092395
- DOI: 10.1371/journal.pone.0001261
Determinants of RNA quality from FFPE samples
Abstract
The large archives of formalin-fixed paraffin-embedded (FFPE) tissue specimens that exist are a highly valuable source of sample material for molecular biological analysis, including gene expression profiling. However, current data on adverse effects of standard pathological practice on the usefulness of biomolecular analytes obtained from such archived specimens is largely anecdotal. Here, we present a systematic examination of the most relevant parameters for integrity and useability of RNA obtained from FFPE samples, including storage time and conditions, fixation time, and specimen size. The results are particularly relevant for any application relying on cDNA synthesis as an initial step of the procedure, such as RT-PCR, and microarray analysis.
Conflict of interest statement
Figures



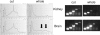
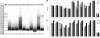
References
-
- Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive–gene expression in paraffin-embedded tissue. J. Pathol. 2001;195:66–71. - PubMed
-
- Bresters D, Schipper MEI, Reesink HW, Boeser-Nunnink BDM, Cuypers HTM. The Duration of Fixation Influences the Yield of HCV cDNA-PCR Products from Formalin-Fixed, Paraffin-Embedded Tissue. J. Virol. Methods. 1994;48:267–272. - PubMed
-
- Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, et al. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod. Pathol. 2002;15:979–987. - PubMed
-
- Bhudevi B, Weinstock D. Detection of Bovine Viral Diarrhea Virus in Formalin Fixed Paraffin embedded Tissue Sections by Real Time RT-PCR. J. Virol. Methods. 2003;109:25–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

