Expression of pyruvate carboxylase mRNA variants in liver of dairy cattle at calving
- PMID: 18060064
- PMCID: PMC2093995
- DOI: 10.1371/journal.pone.0001270
Expression of pyruvate carboxylase mRNA variants in liver of dairy cattle at calving
Abstract
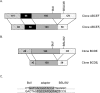
Background: Bovine liver expresses six pyruvate carboxylase (PC) transcript variants, bPC5'A, bPC5'B, bPC5'C, bPC5'D, bPC5'E, and bPC5'F, which only differ at the 5' untranslated region (UTR) and contain a common coding region. The objective of this experiment was to determine the profile and abundance of PC transcripts in bovine liver at calving.
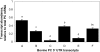
Methodology/principal findings: A ribonuclease protection assay (RPA) protocol was developed to simplify analysis of these variants and investigate the changes in abundance of each 5' UTR transcript relative to total PC mRNA. Liver biopsy samples collected from seven cows on +1 d relative to calving were analyzed by RPA to determine the profile in PC 5' UTR variants. Results show that all six bovine PC 5' UTR variants are detected at calving. Data indicate that bovine PC 5' UTR variant A is the most abundant, variants C and E are least abundant and expression of variants B, D and F is intermediate at calving.
Conclusions: This manuscript describes a simplified RPA method that quantifies the abundance of six PC transcripts by using two riboprobes. The lack of uniformity in the pattern of PC 5' UTR variants at calving suggests an additional complexity for control of bovine PC mRNA expression at calving that may be the result of transcriptional controls, variation in mRNA processing, or a combination of these processes.
Conflict of interest statement
Figures


Similar articles
-
Cloning of bovine pyruvate carboxylase and 5' untranslated region variants.Anim Biotechnol. 2004 May;15(1):47-66. doi: 10.1081/ABIO-120037897. Anim Biotechnol. 2004. PMID: 15248600
-
Bovine pyruvate carboxylase 5' untranslated region variant expression during transition to lactation and feed restriction in dairy cows.J Anim Sci. 2011 Jun;89(6):1881-92. doi: 10.2527/jas.2010-3697. Epub 2011 Jan 28. J Anim Sci. 2011. PMID: 21278109
-
Translational efficiency of bovine pyruvate carboxylase 5' untranslated region messenger ribonucleic acid variants.J Anim Sci. 2008 Dec;86(12):3401-8. doi: 10.2527/jas.2007-0798. Epub 2008 Jul 3. J Anim Sci. 2008. PMID: 18599672
-
Cloning the genomic sequence and identification of promoter regions of bovine pyruvate carboxylase.J Dairy Sci. 2008 Jan;91(1):91-9. doi: 10.3168/jds.2007-0542. J Dairy Sci. 2008. PMID: 18096929
-
Managing the dairy cow at calving time.Vet Clin North Am Food Anim Pract. 2004 Nov;20(3):521-46. doi: 10.1016/j.cvfa.2004.06.001. Vet Clin North Am Food Anim Pract. 2004. PMID: 15471623 Review.
References
-
- Greenfield RB, Cecava MJ, Donkin SS. Changes in mRNA expression for gluconeogenic enzymes in liver of dairy cattle during the transition to lactation. J Dairy Sci. 2000;83:1228–1236. - PubMed
-
- Hartwell JR, Cecava MJ, Donkin SS. Rumen undegradable protein, rumen-protected choline and mRNA expression for enzymes in gluconeogenesis and ureagenesis in periparturient dairy cows. J Dairy Sci. 2001;84:490–497. - PubMed
-
- Velez JC, Donkin SS. Feed restriction induces pyruvate carboxylase but not phosphoenolpyruvate carboxykinase in dairy cows. J Dairy Sci. 2005;88:2938–2948. - PubMed
-
- Bradford BJ, Allen MS. Phlorizin administration increases hepatic gluconeogenic enzyme mRNA abundance but not feed intake in late-lactation dairy cows. J Nutr. 2005;135:2206–2211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

