Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi
- PMID: 18060071
- PMCID: PMC2099475
- DOI: 10.1371/journal.pone.0001277
Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi
Abstract
Background: Microsporidia are intracellular parasites that are highly-derived relatives of fungi. They have compacted genomes and, despite a high rate of sequence evolution, distantly related species can share high levels of gene order conservation. To date, only two species have been analysed in detail, and data from one of these largely consists of short genomic fragments. It is therefore difficult to determine how conservation has been maintained through microsporidian evolution, and impossible to identify whether certain regions are more prone to genomic stasis.
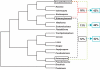
Principal findings: Here, we analyse three large fragments of the Enterocytozoon bieneusi genome (in total 429 kbp), a species of medical significance. A total of 296 ORFs were identified, annotated and their context compared with Encephalitozoon cuniculi and Antonospora locustae. Overall, a high degree of conservation was found between all three species, and interestingly the level of conservation was similar in all three pairwise comparisons, despite the fact that A. locustae is more distantly related to E. cuniculi and E. bieneusi than either are to each other.
Conclusions/significance: Any two genes that are found together in any pair of genomes are more likely to be conserved in the third genome as well, suggesting that a core of genes tends to be conserved across the entire group. The mechanisms of rearrangments identified among microsporidian genomes were consistent with a very slow evolution of their architecture, as opposed to the very rapid sequence evolution reported for these parasites.
Conflict of interest statement
Figures



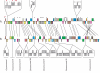
References
-
- Larsson JIR. Identification of microsporidia. Acta Protozoologica. 1999;38:161–197.
-
- Franzen C, Muller A. Microsporidiosis: human diseases and diagnosis. Microbes Infect. 2001;3:389–400. - PubMed
-
- Keeling PJ. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet Biol. 2003;38:298–309. - PubMed
-
- Keeling PJ, Doolittle WF. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol Biol Evol. 1996;13:1297–1305. - PubMed
-
- Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol. 2000;17:23–31. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

