Host gene expression profiling of dengue virus infection in cell lines and patients
- PMID: 18060089
- PMCID: PMC2100376
- DOI: 10.1371/journal.pntd.0000086
Host gene expression profiling of dengue virus infection in cell lines and patients
Abstract
Background: Despite the seriousness of dengue-related disease, with an estimated 50-100 million cases of dengue fever and 250,000-500,000 cases of dengue hemorrhagic fever/dengue shock syndrome each year, a clear understanding of dengue pathogenesis remains elusive. Because of the lack of a disease model in animals and the complex immune interaction in dengue infection, the study of host response and immunopathogenesis is difficult. The development of genomics technology, microarray and high throughput quantitative PCR have allowed researchers to study gene expression changes on a much broader scale. We therefore used this approach to investigate the host response in dengue virus-infected cell lines and in patients developing dengue fever.
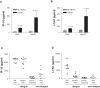
Methodology/principal findings: Using microarray and high throughput quantitative PCR method to monitor the host response to dengue viral replication in cell line infection models and in dengue patient blood samples, we identified differentially expressed genes along three major pathways; NF-kappaB initiated immune responses, type I interferon (IFN) and the ubiquitin proteasome pathway. Among the most highly upregulated genes were the chemokines IP-10 and I-TAC, both ligands of the CXCR3 receptor. Increased expression of IP-10 and I-TAC in the peripheral blood of ten patients at the early onset of fever was confirmed by ELISA. A highly upregulated gene in the IFN pathway, viperin, was overexpressed in A549 cells resulting in a significant reduction in viral replication. The upregulation of genes in the ubiquitin-proteasome pathway prompted the testing of proteasome inhibitors MG-132 and ALLN, both of which reduced viral replication.
Conclusion/significance: Unbiased gene expression analysis has identified new host genes associated with dengue infection, which we have validated in functional studies. We showed that some parts of the host response can be used as potential biomarkers for the disease while others can be used to control dengue viral replication, thus representing viable targets for drug therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Rigau-Perez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, et al. Dengue and dengue haemorrhagic fever. Lancet. 1998;352:971–977. - PubMed
-
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. - PubMed
-
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. - PubMed
-
- Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, et al. Human skin Langerhans cells are targets of dengue virus infection. Nat Med. 2000;6:816–820. - PubMed
-
- Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189:1411–1418. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

