Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity
- PMID: 18061646
- PMCID: PMC2680148
- DOI: 10.1016/j.mad.2007.10.011
Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity
Abstract
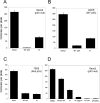
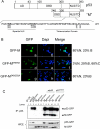
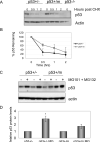
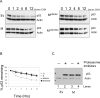
Evidence has accumulated that p53, a prototypical tumor suppressor, may also influence aspects of organismal aging. We have previously described a p53 mutant mouse model, the p53+/m mouse, which is cancer resistant yet exhibits reduced longevity and premature aging phenotypes. p53+/m mice express one full length p53 allele and one truncated p53 allele that is translated into a C-terminal fragment of p53 termed the M protein. The augmented cancer resistance and premature aging phenotypes in the p53+/m mice are consistent with a hyperactive p53 state. To determine how the M protein could increase p53 activity, we examined the M protein in various cellular contexts. Here, we show that embryo fibroblasts from p53+/m mice exhibit reduced proliferation and cell cycle progression compared to embryo fibroblasts from p53+/- mice (with equivalent wild-type p53 dosage). The M protein interacts with wild-type p53, increases its stability, and facilitates its nuclear localization in the absence of stress. Despite increasing p53 stability, the M protein does not disrupt p53-Mdm2 interactions and does not prevent p53 ubiquitination. These results suggest molecular mechanisms by which the M protein could influence the aging and cancer resistance phenotypes in the p53+/m mouse.
Figures




Similar articles
-
Relevance of the p53-MDM2 axis to aging.Cell Death Differ. 2018 Jan;25(1):169-179. doi: 10.1038/cdd.2017.187. Epub 2017 Dec 1. Cell Death Differ. 2018. PMID: 29192902 Free PMC article. Review.
-
Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination.J Biol Chem. 2007 May 11;282(19):14616-25. doi: 10.1074/jbc.M610515200. Epub 2007 Mar 19. J Biol Chem. 2007. PMID: 17371868
-
MDM2 binding induces a conformational change in p53 that is opposed by heat-shock protein 90 and precedes p53 proteasomal degradation.J Biol Chem. 2007 May 11;282(19):14626-34. doi: 10.1074/jbc.M610514200. Epub 2007 Mar 15. J Biol Chem. 2007. PMID: 17363365
-
Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase.Oncogene. 2015 Jan 15;34(3):281-9. doi: 10.1038/onc.2013.557. Epub 2014 Jan 13. Oncogene. 2015. PMID: 24413081
-
Insights into aging obtained from p53 mutant mouse models.Ann N Y Acad Sci. 2004 Jun;1019:171-7. doi: 10.1196/annals.1297.027. Ann N Y Acad Sci. 2004. PMID: 15247009 Review.
Cited by
-
mTORC1 and p53: clash of the gods?Cell Cycle. 2013 Jan 1;12(1):20-5. doi: 10.4161/cc.22912. Cell Cycle. 2013. PMID: 23255104 Free PMC article.
-
Drosophila foxo acts in males to cause sexual-dimorphism in tissue-specific p53 life span effects.Exp Gerontol. 2010 Feb;45(2):97-105. doi: 10.1016/j.exger.2009.10.009. Epub 2009 Oct 17. Exp Gerontol. 2010. PMID: 19840842 Free PMC article.
-
Relevance of the p53-MDM2 axis to aging.Cell Death Differ. 2018 Jan;25(1):169-179. doi: 10.1038/cdd.2017.187. Epub 2017 Dec 1. Cell Death Differ. 2018. PMID: 29192902 Free PMC article. Review.
-
Altered senescence, apoptosis, and DNA damage response in a mutant p53 model of accelerated aging.Mech Ageing Dev. 2009 Apr;130(4):262-71. doi: 10.1016/j.mad.2009.01.001. Mech Ageing Dev. 2009. PMID: 19396980 Free PMC article.
-
The Guardian of the Genome Revisited: p53 Downregulates Genes Required for Telomere Maintenance, DNA Repair, and Centromere Structure.Cancers (Basel). 2018 May 6;10(5):135. doi: 10.3390/cancers10050135. Cancers (Basel). 2018. PMID: 29734785 Free PMC article. Review.
References
-
- Alarcon-Vargas D, Ronai Z. p53-Mdm2--the affair that never ends. Carcinogenesis. 2002;23(4):541–547. - PubMed
-
- Bertrand P, Rouillard D, Boulet A, Levalois C, Soussi T, Lopez BS. Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene. 1997;14(9):1117–1122. - PubMed
-
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

