Use of a multi-thermal washer for DNA microarrays simplifies probe design and gives robust genotyping assays
- PMID: 18063568
- PMCID: PMC2241873
- DOI: 10.1093/nar/gkm1081
Use of a multi-thermal washer for DNA microarrays simplifies probe design and gives robust genotyping assays
Abstract
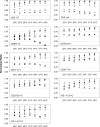
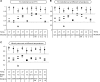
DNA microarrays are generally operated at a single condition, which severely limits the freedom of designing probes for allele-specific hybridization assays. Here, we demonstrate a fluidic device for multi-stringency posthybridization washing of microarrays on microscope slides. This device is called a multi-thermal array washer (MTAW), and it has eight individually controlled heating zones, each of which corresponds to the location of a subarray on a slide. Allele-specific oligonucleotide probes for nine mutations in the beta-globin gene were spotted in eight identical subarrays at positions corresponding to the temperature zones of the MTAW. After hybridization with amplified patient material, the slides were mounted in the MTAW, and each subarray was exposed to different temperatures ranging from 22 to 40 degrees C. When processed in the MTAW, probes selected without considering melting temperature resulted in improved genotyping compared with probes selected according to theoretical melting temperature and run under one condition. In conclusion, the MTAW is a versatile tool that can facilitate screening of a large number of probes for genotyping assays and can also enhance the performance of diagnostic arrays.
Figures



Similar articles
-
Microfludic device for creating ionic strength gradients over DNA microarrays for efficient DNA melting studies and assay development.PLoS One. 2009;4(3):e4808. doi: 10.1371/journal.pone.0004808. Epub 2009 Mar 11. PLoS One. 2009. PMID: 19277213 Free PMC article.
-
Solution-phase DNA mutation scanning and SNP genotyping by nanoliter melting analysis.Biomed Microdevices. 2007 Apr;9(2):159-66. doi: 10.1007/s10544-006-9017-3. Biomed Microdevices. 2007. PMID: 17165128
-
Detection of mutations using microarrays of poly(C)10-poly(T)10 modified DNA probes immobilized on agarose films.Anal Biochem. 2006 May 15;352(2):188-97. doi: 10.1016/j.ab.2006.03.008. Epub 2006 Mar 23. Anal Biochem. 2006. PMID: 16615930
-
DNA microarrays.Adv Biochem Eng Biotechnol. 2008;109:433-53. doi: 10.1007/10_2007_087. Adv Biochem Eng Biotechnol. 2008. PMID: 17985099 Review.
-
Single nucleotide polymorphism genotyping using locked nucleic acid (LNA).Expert Rev Mol Diagn. 2003 Jan;3(1):27-38. doi: 10.1586/14737159.3.1.27. Expert Rev Mol Diagn. 2003. PMID: 12528362 Review.
Cited by
-
A Biochemical Corrosion Monitoring Sensor with a Silver/Carbon Comb Structure for the Detection of Living Escherichia coli.ACS Omega. 2023 Nov 13;8(46):43511-43520. doi: 10.1021/acsomega.3c03632. eCollection 2023 Nov 21. ACS Omega. 2023. PMID: 38027348 Free PMC article.
-
Hybridization thermodynamics of NimbleGen microarrays.BMC Bioinformatics. 2010 Jan 19;11:35. doi: 10.1186/1471-2105-11-35. BMC Bioinformatics. 2010. PMID: 20085625 Free PMC article.
-
SPR Detection and Discrimination of the Oligonucleotides Related to the Normal and the Hybrid bcr-abl Genes by Two Stringency Control Strategies.Nanoscale Res Lett. 2016 Dec;11(1):19. doi: 10.1186/s11671-016-1226-y. Epub 2016 Jan 13. Nanoscale Res Lett. 2016. PMID: 26759355 Free PMC article.
-
Prediction of the optimum hybridization conditions of dot-blot-SNP analysis using estimated melting temperature of oligonucleotide probes.Plant Cell Rep. 2010 Aug;29(8):829-34. doi: 10.1007/s00299-010-0867-z. Epub 2010 May 21. Plant Cell Rep. 2010. PMID: 20490503
-
Investigation of parameters that affect the success rate of microarray-based allele-specific hybridization assays.PLoS One. 2011 Mar 22;6(3):e14777. doi: 10.1371/journal.pone.0014777. PLoS One. 2011. PMID: 21445337 Free PMC article.
References
-
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. - PubMed
-
- Syvanen AC. Toward genome-wide SNP genotyping. Nat. Genet. 2005;37(Suppl.):S5–10. - PubMed
-
- Vainrub A, Pettitt BM. Theoretical aspects of genomic variation screening using DNA microarrays. Biopolymers. 2004;73:614–620. - PubMed
-
- Vainrub A, Pettitt BM. Surface electrostatic effects in oligonucleotide microarrays: control and optimization of binding thermodynamics. Biopolymers. 2003;68:265–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

