PlantGDB: a resource for comparative plant genomics
- PMID: 18063570
- PMCID: PMC2238959
- DOI: 10.1093/nar/gkm1041
PlantGDB: a resource for comparative plant genomics
Abstract
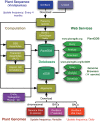
PlantGDB (http://www.plantgdb.org/) is a genomics database encompassing sequence data for green plants (Viridiplantae). PlantGDB provides annotated transcript assemblies for >100 plant species, with transcripts mapped to their cognate genomic context where available, integrated with a variety of sequence analysis tools and web services. For 14 plant species with emerging or complete genome sequence, PlantGDB's genome browsers (xGDB) serve as a graphical interface for viewing, evaluating and annotating transcript and protein alignments to chromosome or bacterial artificial chromosome (BAC)-based genome assemblies. Annotation is facilitated by the integrated yrGATE module for community curation of gene models. Novel web services at PlantGDB include Tracembler, an iterative alignment tool that generates contigs from GenBank trace file data and BioExtract Server, a web-based server for executing custom sequence analysis workflows. PlantGDB also hosts a plant genomics research outreach portal (PGROP) that facilitates access to a large number of resources for research and training.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

