Mesaconyl-coenzyme A hydratase, a new enzyme of two central carbon metabolic pathways in bacteria
- PMID: 18065535
- PMCID: PMC2238226
- DOI: 10.1128/JB.01621-07
Mesaconyl-coenzyme A hydratase, a new enzyme of two central carbon metabolic pathways in bacteria
Abstract
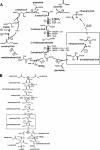
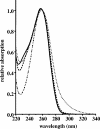
The coenzyme A (CoA)-activated C5-dicarboxylic acids mesaconyl-CoA and beta-methylmalyl-CoA play roles in two as yet not completely resolved central carbon metabolic pathways in bacteria. First, these compounds are intermediates in the 3-hydroxypropionate cycle for autotrophic CO2 fixation in Chloroflexus aurantiacus, a phototrophic green nonsulfur bacterium. Second, mesaconyl-CoA and beta-methylmalyl-CoA are intermediates in the ethylmalonyl-CoA pathway for acetate assimilation in various bacteria, e.g., in Rhodobacter sphaeroides, Methylobacterium extorquens, and Streptomyces species. In both cases, mesaconyl-CoA hydratase was postulated to catalyze the interconversion of mesaconyl-CoA and beta-methylmalyl-CoA. The putative genes coding for this enzyme in C. aurantiacus and R. sphaeroides were cloned and heterologously expressed in Escherichia coli, and the proteins were purified and studied. The recombinant homodimeric 80-kDa proteins catalyzed the reversible dehydration of erythro-beta-methylmalyl-CoA to mesaconyl-CoA with rates of 1,300 micromol min(-1) mg protein(-1). Genes coding for similar enzymes with two (R)-enoyl-CoA hydratase domains are present in the genomes of Roseiflexus, Methylobacterium, Hyphomonas, Rhodospirillum, Xanthobacter, Caulobacter, Magnetospirillum, Jannaschia, Sagittula, Parvibaculum, Stappia, Oceanicola, Loktanella, Silicibacter, Roseobacter, Roseovarius, Dinoroseobacter, Sulfitobacter, Paracoccus, and Ralstonia species. A similar yet distinct class of enzymes containing only one hydratase domain was found in various other bacteria, such as Streptomyces species. The role of this widely distributed new enzyme is discussed.
Figures



References
-
- Alber, B. E., and G. Fuchs. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 27712137-12143. - PubMed
-
- Alber, B. E., R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 61297-309. - PubMed
-
- Albers, H., and G. Gottschalk. 1976. Acetate metabolism in Rhodopseudomonas gelatinosa and several other Rhodospirillaceae. Arch. Microbiol. 11145-49. - PubMed
-
- Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

