Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus
- PMID: 18065539
- PMCID: PMC2238196
- DOI: 10.1128/JB.01415-07
Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus
Abstract
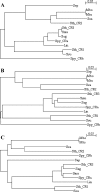
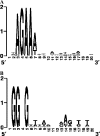
Clustered regularly interspaced short palindromic repeats (CRISPR) are hypervariable loci widely distributed in prokaryotes that provide acquired immunity against foreign genetic elements. Here, we characterize a novel Streptococcus thermophilus locus, CRISPR3, and experimentally demonstrate its ability to integrate novel spacers in response to bacteriophage. Also, we analyze CRISPR diversity and activity across three distinct CRISPR loci in several S. thermophilus strains. We show that both CRISPR repeats and cas genes are locus specific and functionally coupled. A total of 124 strains were studied, and 109 unique spacer arrangements were observed across the three CRISPR loci. Overall, 3,626 spacers were analyzed, including 2,829 for CRISPR1 (782 unique), 173 for CRISPR2 (16 unique), and 624 for CRISPR3 (154 unique). Sequence analysis of the spacers revealed homology and identity to phage sequences (77%), plasmid sequences (16%), and S. thermophilus chromosomal sequences (7%). Polymorphisms were observed for the CRISPR repeats, CRISPR spacers, cas genes, CRISPR motif, locus architecture, and specific sequence content. Interestingly, CRISPR loci evolved both via polarized addition of novel spacers after exposure to foreign genetic elements and via internal deletion of spacers. We hypothesize that the level of diversity is correlated with relative CRISPR activity and propose that the activity is highest for CRISPR1, followed by CRISPR3, while CRISPR2 may be degenerate. Globally, the dynamic nature of CRISPR loci might prove valuable for typing and comparative analyses of strains and microbial populations. Also, CRISPRs provide critical insights into the relationships between prokaryotes and their environments, notably the coevolution of host and viral genomes.
Figures
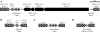




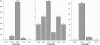
References
-
- Barrangou, R., C. Fremaux, P. Boyaval, M. Richards, H. Deveau, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. - PubMed
-
- Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyrpides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 221554-1558. - PMC - PubMed
-
- Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 1512551-2561. - PubMed
-
- Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H. P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 2731058-1073. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

