Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella
- PMID: 18065549
- PMCID: PMC2293233
- DOI: 10.1128/JB.01620-07
Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella
Abstract
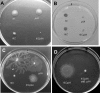
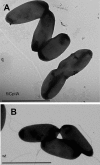
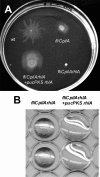
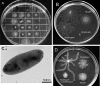
Pseudomonas aeruginosa exhibits swarming motility on 0.5 to 1% agar plates in the presence of specific carbon and nitrogen sources. We have found that PAO1 double mutants expressing neither flagella nor type IV pili (fliC pilA) display sliding motility under the same conditions. Sliding motility was inhibited when type IV pilus expression was restored; like swarming motility, it also decreased in the absence of rhamnolipid surfactant production. Transposon insertions in gacA and gacS increased sliding motility and restored tendril formation to spreading colonies, while transposon insertions in retS abolished motility. These changes in motility were not accompanied by detectable changes in rhamnolipid surfactant production or by the appearance of bacterial surface structures that might power sliding motility. We propose that P. aeruginosa requires flagella during swarming to overcome adhesive interactions mediated by type IV pili. The apparent dependence of sliding motility on environmental cues and regulatory pathways that also affect swarming motility suggests that both forms of motility are influenced by similar cohesive factors that restrict translocation, as well as by dispersive factors that facilitate spreading. Studies of sliding motility may be particularly well-suited for identifying factors other than pili and flagella that affect community behaviors of P. aeruginosa.
Figures
Similar articles
-
The Pseudomonas aeruginosa PilSR Two-Component System Regulates Both Twitching and Swimming Motilities.mBio. 2018 Jul 24;9(4):e01310-18. doi: 10.1128/mBio.01310-18. mBio. 2018. PMID: 30042200 Free PMC article.
-
Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili.J Bacteriol. 2000 Nov;182(21):5990-6. doi: 10.1128/JB.182.21.5990-5996.2000. J Bacteriol. 2000. PMID: 11029417 Free PMC article.
-
Swarming of Pseudomonas aeruginosa PAO1 without differentiation into elongated hyperflagellates on hard agar minimal medium.FEMS Microbiol Lett. 2008 Mar;280(2):169-75. doi: 10.1111/j.1574-6968.2007.01057.x. Epub 2008 Jan 31. FEMS Microbiol Lett. 2008. PMID: 18248427
-
Cross-regulation of Pseudomonas motility systems: the intimate relationship between flagella, pili and virulence.Curr Opin Microbiol. 2015 Dec;28:78-82. doi: 10.1016/j.mib.2015.07.017. Epub 2015 Oct 23. Curr Opin Microbiol. 2015. PMID: 26476804 Free PMC article. Review.
-
The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa--a review.Gene. 1996 Nov 7;179(1):147-55. doi: 10.1016/s0378-1119(96)00441-6. Gene. 1996. PMID: 8955641 Review.
Cited by
-
Exopolysaccharides amylovoran and levan contribute to sliding motility in the fire blight pathogen Erwinia amylovora.Environ Microbiol. 2022 Oct;24(10):4738-4754. doi: 10.1111/1462-2920.16193. Epub 2022 Sep 15. Environ Microbiol. 2022. PMID: 36054324 Free PMC article.
-
A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence.PLoS One. 2009;4(2):e4358. doi: 10.1371/journal.pone.0004358. Epub 2009 Feb 4. PLoS One. 2009. PMID: 19194503 Free PMC article.
-
Swarming of P. aeruginosa: Through the lens of biophysics.Biophys Rev (Melville). 2023 Sep;4(3):031305. doi: 10.1063/5.0128140. Epub 2023 Sep 28. Biophys Rev (Melville). 2023. PMID: 37781002 Free PMC article. Review.
-
Bacterial motility: machinery and mechanisms.Nat Rev Microbiol. 2022 Mar;20(3):161-173. doi: 10.1038/s41579-021-00626-4. Epub 2021 Sep 21. Nat Rev Microbiol. 2022. PMID: 34548639 Review.
-
Genotypic and phenotypic analyses of two distinct sets of Pseudomonas aeruginosa urinary tract isolates.J Med Microbiol. 2025 Feb;74(2):001971. doi: 10.1099/jmm.0.001971. J Med Microbiol. 2025. PMID: 40013918 Free PMC article.
References
-
- Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single copy chromosomal lacZ and lux gene fusions. BioTechniques 29948-952. - PubMed
-
- Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64391-397. - PubMed
-
- Dasgupta, N., M. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

