Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice
- PMID: 18065552
- PMCID: PMC2245831
- DOI: 10.1104/pp.107.112821
Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice
Abstract
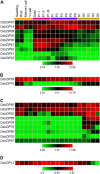
The basic leucine (Leu) zipper (bZIP) proteins compose a family of transcriptional regulators present exclusively in eukaryotes. The bZIP proteins characteristically harbor a bZIP domain composed of two structural features: a DNA-binding basic region and the Leu zipper dimerization region. They have been shown to regulate diverse plant-specific phenomena, including seed maturation and germination, floral induction and development, and photomorphogenesis, and are also involved in stress and hormone signaling. We have identified 89 bZIP transcription factor-encoding genes in the rice (Oryza sativa) genome. Their chromosomal distribution and sequence analyses suggest that the bZIP transcription factor family has evolved via gene duplication. The phylogenetic relationship among rice bZIP domains as well as with bZIP domains from other plant bZIP factors suggests that homologous bZIP domains exist in plants. Similar intron/exon structural patterns were observed in the basic and hinge regions of their bZIP domains. Detailed sequence analysis has been done to identify additional conserved motifs outside the bZIP domain and to predict their DNA-binding site specificity as well as dimerization properties, which has helped classify them into different groups and subfamilies, respectively. Expression of bZIP transcription factor-encoding genes has been analyzed by full-length cDNA and expressed sequence tag-based expression profiling. This expression profiling was complemented by microarray analysis. The results indicate specific or coexpression patterns of rice bZIP transcription factors starting from floral transition to various stages of panicle and seed development. bZIP transcription factor-encoding genes in rice also displayed differential expression patterns in rice seedlings in response to abiotic stress and light irradiation. An effort has been made to link the structure and expression pattern of bZIP transcription factor-encoding genes in rice to their function, based on the information obtained from our analyses and earlier known results. This information will be important for functional characterization of bZIP transcription factors in rice.
Figures



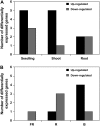

References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056 - PubMed
-
- Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C (2002) A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry 41 14122–14131 - PubMed
-
- Aeschbacher RA, Schrott M, Potrykus I, Saul MW (1991) Isolation and molecular characterization of PosF21, an Arabidopsis thaliana gene which shows characteristics of a b-Zip class transcription factor. Plant J 1 303–316 - PubMed
-
- Aguan K, Sugawara K, Suzuki N, Kusano T (1993) Low-temperature-dependent expression of a rice gene encoding a protein with a leucine-zipper motif. Mol Gen Genet 240 1–8 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

