Firefly luciferase complementation imaging assay for protein-protein interactions in plants
- PMID: 18065554
- PMCID: PMC2245818
- DOI: 10.1104/pp.107.111740
Firefly luciferase complementation imaging assay for protein-protein interactions in plants
Abstract
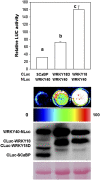
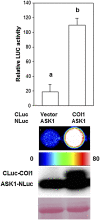
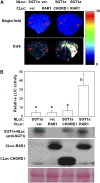
The development of sensitive and versatile techniques to detect protein-protein interactions in vivo is important for understanding protein functions. The previously described techniques, fluorescence resonance energy transfer and bimolecular fluorescence complementation, which are used widely for protein-protein interaction studies in plants, require extensive instrumentation. To facilitate protein-protein interaction studies in plants, we adopted the luciferase complementation imaging assay. The amino-terminal and carboxyl-terminal halves of the firefly luciferase reconstitute active luciferase enzyme only when fused to two interacting proteins, and that can be visualized with a low-light imaging system. A series of plasmid constructs were made to enable the transient expression of fusion proteins or generation of stable transgenic plants. We tested nine pairs of proteins known to interact in plants, including Pseudomonas syringae bacterial effector proteins and their protein targets in the plant, proteins of the SKP1-Cullin-F-box protein E3 ligase complex, the HSP90 chaperone complex, components of disease resistance protein complex, and transcription factors. In each case, strong luciferase complementation was observed for positive interactions. Mutants that are known to compromise protein-protein interactions showed little or much reduced luciferase activity. Thus, the assay is simple, reliable, and quantitative in detection of protein-protein interactions in plants.
Figures


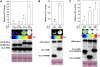
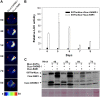
References
-
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2077–2080 - PubMed
-
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40 419–427 - PubMed
-
- Callis J, Vierstra RD (2000) Protein degradation in signaling. Curr Opin Plant Biol 3 381–386 - PubMed
-
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

