Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus
- PMID: 18065566
- PMCID: PMC2245823
- DOI: 10.1104/pp.107.107060
Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus
Abstract
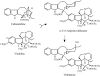
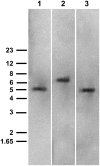
Catharanthus roseus produces low levels of two dimeric terpenoid indole alkaloids, vinblastine and vincristine, which are widely used in cancer chemotherapy. The dimerization reaction leading to alpha-3',4'-anhydrovinblastine is a key regulatory step for the production of the anticancer alkaloids in planta and has potential application in the industrial production of two semisynthetic derivatives also used as anticancer drugs. In this work, we report the cloning, characterization, and subcellular localization of an enzyme with anhydrovinblastine synthase activity identified as the major class III peroxidase present in C. roseus leaves and named CrPrx1. The deduced amino acid sequence corresponds to a polypeptide of 363 amino acids including an N-terminal signal peptide showing the secretory nature of CrPrx1. CrPrx1 has a two-intron structure and is present as a single gene copy. Phylogenetic analysis indicates that CrPrx1 belongs to an evolutionary branch of vacuolar class III peroxidases whose members seem to have been recruited for different functions during evolution. Expression of a green fluorescent protein-CrPrx1 fusion confirmed the vacuolar localization of this peroxidase, the exact subcellular localization of the alkaloid monomeric precursors and dimeric products. Expression data further supports the role of CrPrx1 in alpha-3',4'-anhydrovinblastine biosynthesis, indicating the potential of CrPrx1 as a target to increase alkaloid levels in the plant.
Figures






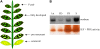


Similar articles
-
Cloning, characterization and localization of a novel basic peroxidase gene from Catharanthus roseus.FEBS J. 2007 Mar;274(5):1290-303. doi: 10.1111/j.1742-4658.2007.05677.x. FEBS J. 2007. PMID: 17298442
-
A vacuolar class III peroxidase and the metabolism of anticancer indole alkaloids in Catharanthus roseus: Can peroxidases, secondary metabolites and arabinogalactan proteins be partners in microcompartmentation of cellular reactions?Plant Signal Behav. 2008 Oct;3(10):899-901. doi: 10.4161/psb.3.10.6576. Plant Signal Behav. 2008. PMID: 19704535 Free PMC article.
-
Ectopic overexpression of vacuolar and apoplastic Catharanthus roseus peroxidases confers differential tolerance to salt and dehydration stress in transgenic tobacco.Protoplasma. 2012 Apr;249(2):423-32. doi: 10.1007/s00709-011-0294-1. Epub 2011 Jun 4. Protoplasma. 2012. PMID: 21643888
-
Enzymology of indole alkaloid biosynthesis in Catharanthus roseus.Indian J Biochem Biophys. 1996 Aug;33(4):261-73. Indian J Biochem Biophys. 1996. PMID: 8936815 Review.
-
[Advance in biosynthesis of terpenoid indole alkaloids and its regulation in Catharanthus roseus].Zhongguo Zhong Yao Za Zhi. 2016 Nov;41(22):4129-4137. doi: 10.4268/cjcmm20162208. Zhongguo Zhong Yao Za Zhi. 2016. PMID: 28933078 Review. Chinese.
Cited by
-
Towards understanding vacuolar antioxidant mechanisms: a role for fructans?J Exp Bot. 2013 Feb;64(4):1025-38. doi: 10.1093/jxb/ers377. Epub 2013 Jan 23. J Exp Bot. 2013. PMID: 23349141 Free PMC article.
-
Repression of ZCT1, ZCT2 and ZCT3 affects expression of terpenoid indole alkaloid biosynthetic and regulatory genes.PeerJ. 2021 Jul 2;9:e11624. doi: 10.7717/peerj.11624. eCollection 2021. PeerJ. 2021. PMID: 34249496 Free PMC article.
-
Characterization of a second secologanin synthase isoform producing both secologanin and secoxyloganin allows enhanced de novo assembly of a Catharanthus roseus transcriptome.BMC Genomics. 2015 Aug 19;16(1):619. doi: 10.1186/s12864-015-1678-y. BMC Genomics. 2015. PMID: 26285573 Free PMC article.
-
Folivory elicits a strong defense reaction in Catharanthus roseus: metabolomic and transcriptomic analyses reveal distinct local and systemic responses.Sci Rep. 2017 Jan 17;7:40453. doi: 10.1038/srep40453. Sci Rep. 2017. PMID: 28094274 Free PMC article.
-
Molecular cloning and characterization of seven class III peroxidases induced by overexpression of the agrobacterial rolB gene in Rubia cordifolia transgenic callus cultures.Plant Cell Rep. 2012 Jun;31(6):1009-19. doi: 10.1007/s00299-011-1219-3. Epub 2012 Jan 12. Plant Cell Rep. 2012. PMID: 22238062
References
-
- Barceló AR, Gómez Ros LV, Gabaldón C, López-Serrano M, Pomar F, Carrión JS, Pedreño MA (2004) Basic peroxidases: the gateway for lignin evolution? Phytochem Rev 3 61–78
-
- Bernards MA, Summerhurst DK, Razem FA (2004) Oxidases, peroxidases and hydrogen peroxide: the suberin connection. Phytochem Rev 3 113–126
-
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6 325–330 - PubMed
-
- Díaz J, Pomar F, Bernal A, Merino F (2004) Peroxidases and the metabolism of capsaicin in Capsicum annuum L. Phytochem Rev 3 141–157
-
- Duroux L, Welinder KG (2003) The peroxidase gene family in plants: a phylogenetic overview. J Mol Evol 57 397–407 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

