Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis
- PMID: 18065688
- PMCID: PMC2217644
- DOI: 10.1105/tpc.107.054312
Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis
Abstract
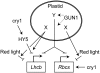
Plastid signals are among the most potent regulators of genes that encode proteins active in photosynthesis. Plastid signals help coordinate the expression of the nuclear and chloroplast genomes and the expression of genes with the functional state of the chloroplast. Here, we report the isolation of new cryptochrome1 (cry1) alleles from a screen for Arabidopsis thaliana genomes uncoupled mutants, which have defects in plastid-to-nucleus signaling. We also report genetic experiments showing that a previously unidentified plastid signal converts multiple light signaling pathways that perceive distinct qualities of light from positive to negative regulators of some but not all photosynthesis-associated nuclear genes (PhANGs) and change the fluence rate response of PhANGs. At least part of this remodeling of light signaling networks involves converting HY5, a positive regulator of PhANGs, into a negative regulator of PhANGs. We also observed that mutants with defects in both plastid-to-nucleus and cry1 signaling exhibited severe chlorophyll deficiencies. These data show that the remodeling of light signaling networks by plastid signals is a mechanism that plants use to integrate signals describing the functional and developmental state of plastids with signals describing particular light environments when regulating PhANG expression and performing chloroplast biogenesis.
Figures






References
-
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166. - PubMed
-
- Ahmad, M., Lin, C., and Cashmore, A.R. (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 8 653–658. - PubMed
-
- Albrecht, V., Ingenfeld, A., and Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60 507–518. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

