Characterization of the Drosophila segment determination morphome
- PMID: 18067886
- PMCID: PMC2254320
- DOI: 10.1016/j.ydbio.2007.10.037
Characterization of the Drosophila segment determination morphome
Abstract
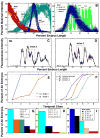
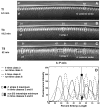
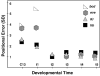
Here we characterize the expression of the full system of genes which control the segmentation morphogenetic field of Drosophila at the protein level in one dimension. The data used for this characterization are quantitative with cellular resolution in space and about 6 min in time. We present the full quantitative profiles of all 14 segmentation genes which act before the onset of gastrulation. The expression patterns of these genes are first characterized in terms of their average or typical behavior. At this level, the expression of all of the genes has been integrated into a single atlas of gene expression in which the expression levels of all genes in each cell are specified. We show that expression domains do not arise synchronously, but rather each domain has its own specific dynamics of formation. Moreover, we show that the expression domains shift position in the direction of the cephalic furrow, such that domains in the anlage of the segmented germ band shift anteriorly while those in the presumptive head shift posteriorly. The expression atlas of integrated data is very close to the expression profiles of individual embryos during the latter part of the blastoderm stage. At earlier times gap gene domains show considerable variation in amplitude, and significant positional variability. Nevertheless, an average early gap domain is close to that of a median individual. In contrast, we show that there is a diversity of developmental trajectories among pair-rule genes at a variety of levels, including the order of domain formation and positional accuracy. We further show that this variation is dynamically reduced, or canalized, over time. As the first quantitatively characterized morphogenetic field, this system and its behavior constitute an extraordinarily rich set of materials for the study of canalization and embryonic regulation at the molecular level.
Figures






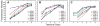

References
-
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. - PubMed
-
- Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. - PubMed
-
- Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8:105–117. - PubMed
-
- Blankenship JT, Wieschaus E. Two new roles for the Drosophila AP patterning system in early morphogenesis. Development. 2001;128:5129–5138. - PubMed
-
- Broomhead D, King G. Extracting qualitative dynamics from experimental data. Physica D. 1986;20:217–236.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

