Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization
- PMID: 18068724
- PMCID: PMC2289799
- DOI: 10.1016/j.jmb.2007.11.013
Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization
Abstract
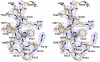
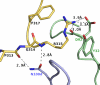
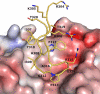
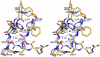
F425-B4e8 (B4e8) is a monoclonal antibody isolated from a human immunodeficiency virus type 1 (HIV-1)-infected individual that recognizes the V3 variable loop on the gp120 subunit of the viral envelope spike. B4e8 neutralizes a subset of HIV-1 primary isolates from subtypes B, C and D, which places this antibody among the very few human anti-V3 antibodies with notable cross-neutralizing activity. Here, the crystal structure of the B4e8 Fab' fragment in complex with a 24-mer V3 peptide (RP142) at 2.8 A resolution is described. The complex structure reveals that the antibody recognizes a novel V3 loop conformation, featuring a five-residue alpha-turn around the conserved GPGRA apex of the beta-hairpin loop. In agreement with previous mutagenesis analyses, the Fab' interacts primarily with V3 through side-chain contacts with just two residues, Ile(P309) and Arg(P315), while the remaining contacts are to the main chain. The structure helps explain how B4e8 can tolerate a certain degree of sequence variation within V3 and, hence, is able to neutralize an appreciable number of different HIV-1 isolates.
Figures
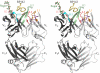
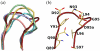
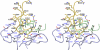
References
-
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. - PubMed
-
- Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. - PubMed
-
- Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 2005;79:1252–1261. - PMC - PubMed
-
- Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

