Comparative sialomics between hard and soft ticks: implications for the evolution of blood-feeding behavior
- PMID: 18070664
- PMCID: PMC2211429
- DOI: 10.1016/j.ibmb.2007.09.003
Comparative sialomics between hard and soft ticks: implications for the evolution of blood-feeding behavior
Abstract
Ticks evolved various mechanisms to modulate their host's hemostatic and immune defenses. Differences in the anti-hemostatic repertoires suggest that hard and soft ticks evolved anti-hemostatic mechanisms independently, but raise questions on the conservation of salivary gland proteins in the ancestral tick lineage. To address this issue, the sialome (salivary gland secretory proteome) from the soft tick, Argas monolakensis, was determined by proteomic analysis and cDNA library construction of salivary glands from fed and unfed adult female ticks. The sialome is composed of approximately 130 secretory proteins of which the most abundant protein folds are the lipocalin, BTSP, BPTI and metalloprotease families which also comprise the most abundant proteins found in the salivary glands. Comparative analysis indicates that the major protein families are conserved in hard and soft ticks. Phylogenetic analysis shows, however, that most gene duplications are lineage specific, indicating that the protein families analyzed possibly evolved most of their functions after divergence of the two major tick families. In conclusion, the ancestral tick may have possessed a simple (few members for each family), but diverse (many different protein families) salivary gland protein domain repertoire.
Figures
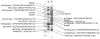



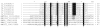


References
-
- Alarcon-Chaidez FJ, Sun J, Wikel SK. Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari: Ixodidae) Insect Biochem Mol Biol. 2007;37:48–71. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST - a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. - PubMed
-
- Barker SC, Murrell A. Systematics and evolution of ticks with a list of valid genus and species names. Parasitology Suppl. 2004;129:15–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

