A non-tumor suppressor role for basal p19ARF in maintaining nucleolar structure and function
- PMID: 18070929
- PMCID: PMC2223401
- DOI: 10.1128/MCB.00484-07
A non-tumor suppressor role for basal p19ARF in maintaining nucleolar structure and function
Abstract
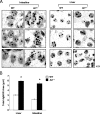
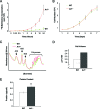
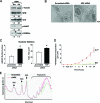
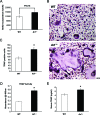
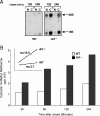
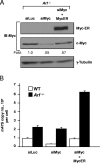
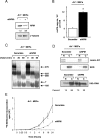
The nucleolus is the center of ribosome synthesis, with the nucleophosmin (NPM) and p19(ARF) proteins antagonizing one another to either promote or inhibit growth. However, basal NPM and ARF proteins form nucleolar complexes whose functions remain unknown. Nucleoli from Arf(-/)(-) cells displayed increased nucleolar area, suggesting that basal ARF might regulate key nucleolar functions. Concordantly, ribosome biogenesis and protein synthesis were dramatically elevated in the absence of Arf, causing these cells to exhibit tremendous gains in protein amounts and increases in cell volume. The transcription of ribosomal DNA (rDNA), the processing of nascent rRNA molecules, and the nuclear export of ribosomes were all increased in the absence of ARF. Similar results were obtained using targeted lentiviral RNA interference of ARF in wild-type MEFs. Postmitotic osteoclasts from Arf-null mice exhibited hyperactivity in vitro and in vivo, demonstrating a physiological function for basal ARF. Moreover, the knockdown of NPM blocked the increases in Arf(-/-) ribosome output and osteoclast activity, demonstrating that these gains require NPM. Thus, basal ARF proteins act as a monitor of steady-state ribosome biogenesis and growth independent of their ability to prevent unwarranted hyperproliferation.
Figures




References
-
- Aubele, M., S. Biesterfeld, M. Derenzini, P. Hufnagl, H. Martin, D. Ofner, D. Ploton, and J. Ruschoff for the Committee on AgNOR Quantitation within the European Society of Pathology. 1994. Guidelines of AgNOR quantitation. Zentralbl. Pathol. 140107-108. - PubMed
-
- Ayrault, O., L. Andrique, D. Fauvin, B. Eymin, S. Gazzeri, and P. Seite. 2006. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 257577-7786. - PubMed
-
- Ayrault, O., L. Andrique, C. J. Larsen, and P. Seite. 2004. Human Arf tumor suppressor specifically interacts with chromatin containing the promoter of rRNA genes. Oncogene 238097-8104. - PubMed
-
- Bertwistle, D., F. Zindy, C. J. Sherr, and M. F. Roussel. 2004. Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybrid. Hybridomics 23293-300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
