Frequent deletion of the CDKN2A locus in chordoma: analysis of chromosomal imbalances using array comparative genomic hybridisation
- PMID: 18071362
- PMCID: PMC2361468
- DOI: 10.1038/sj.bjc.6604130
Frequent deletion of the CDKN2A locus in chordoma: analysis of chromosomal imbalances using array comparative genomic hybridisation
Abstract
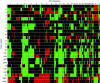
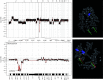
The initiating somatic genetic events in chordoma development have not yet been identified. Most cytogenetically investigated chordomas have displayed near-diploid or moderately hypodiploid karyotypes, with several numerical and structural rearrangements. However, no consistent structural chromosome aberration has been reported. This is the first array-based study characterising DNA copy number changes in chordoma. Array comparative genomic hybridisation (aCGH) identified copy number alterations in all samples and imbalances affecting 5 or more out of the 21 investigated tumours were seen on all chromosomes. In general, deletions were more common than gains and no high-level amplification was found, supporting previous findings of primarily losses of large chromosomal regions as an important mechanism in chordoma development. Although small imbalances were commonly found, the vast majority of these were detected in single cases; no small deletion affecting all tumours could be discerned. However, the CDKN2A and CDKN2B loci in 9p21 were homo- or heterozygously lost in 70% of the tumours, a finding corroborated by fluorescence in situ hybridisation, suggesting that inactivation of these genes constitute an important step in chordoma development.
Figures

References
-
- Autio R, Hautaniemi S, Kauraniemi P, Yli-Harja O, Astola J, Wolf M, Kallioniemi A (2003) CGH-Plotter: MATLAB toolbox for CGH-data analysis. Bioinformatics 19: 1714–1715 - PubMed
-
- Blyth K, Cameron ER, Neil JC (2005) The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5: 376–387 - PubMed
-
- Bovee JVMG, Sciot R, Dal Cin P, Debiec-Rychter M, van Zelderen-Bhola SL, Cornelisse CJ, Hogendoorn PCW (2001) Chromosome 9 alterations and trisomy 22 in central chondrosarcoma: a cytogenetic and DNA flow cytometric analysis of chondrosarcoma subtypes. Diagn Mol Pathol 10: 228–235 - PubMed
-
- Brandal P, Bjerkehagen B, Danielsen H, Heim S (2005) Chromosome 7 abnormalities are common in chordomas. Cancer Genet Cytogenet 160: 15–21 - PubMed
-
- Dahlén A, Debiec-Rychter M, Pedeutour F, Domanski HA, Höglund M, Bauer HCF, Rydholm A, Sciot R, Mandahl N, Mertens F (2003) Clustering of deletions on chromosome 13 in benign and low-malignant lipomatous tumors. Int J Cancer 103: 616–623 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

