Specific RNA self-assembly with minimal paranemic motifs
- PMID: 18072767
- PMCID: PMC4869885
- DOI: 10.1021/ja071516m
Specific RNA self-assembly with minimal paranemic motifs
Abstract
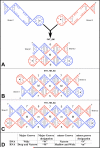
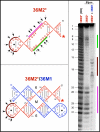
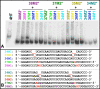
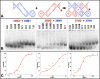
The paranemic crossover (PX) is a motif for assembling two nucleic acid molecules using Watson-Crick (WC) basepairing without unfolding preformed secondary structure in the individual molecules. Once formed, the paranemic assembly motif comprises adjacent parallel double helices that crossover at every possible point over the length of the motif. The interaction is reversible as it does not require denaturation of basepairs internal to each interacting molecular unit. Paranemic assembly has been demonstrated for DNA but not for RNA and only for motifs with four or more crossover points and lengths of five or more helical half-turns. Here we report the design of RNA molecules that paranemically assemble with the minimum number of two crossovers spanning the major groove to form paranemic motifs with a length of three half turns (3HT). Dissociation constants (Kd's) were measured for a series of molecules in which the number of basepairs between the crossover points was varied from five to eight basepairs. The paranemic 3HT complex with six basepairs (3HT_6M) was found to be the most stable with Kd = 1 x 10-8 M. The half-time for kinetic exchange of the 3HT_6M complex was determined to be approximately 100 min, from which we calculated association and dissociation rate constants ka = 5.11 x 103 M-1s-1 and kd = 5.11 x 10-5 s-1. RNA paranemic assembly of 3HT and 5HT complexes is blocked by single-base substitutions that disrupt individual intermolecular Watson-Crick basepairs and is restored by compensatory substitutions that restore those basepairs. The 3HT motif appears suitable for specific, programmable, and reversible tecto-RNA self-assembly for constructing artificial RNA molecular machines.
Figures





References
-
- Ball P. Nanotechnology. 2002;13:R15–R28.
-
- Goodsell DS. Bionanotechnology: Lessons from Nature. Wiley-Liss; Hoboken, New Jersey: 2004.
-
- Jaeger L, Leontis NB. Angew Chem Int Ed Engl. 2000;39:2521–2524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

