Protein evolution by hypermutation and selection in the B cell line DT40
- PMID: 18073192
- PMCID: PMC2248763
- DOI: 10.1093/nar/gkm616
Protein evolution by hypermutation and selection in the B cell line DT40
Abstract
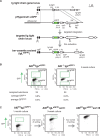
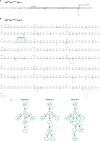
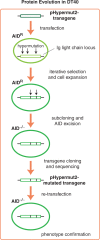
Genome-wide mutations and selection within a population are the basis of natural evolution. A similar process occurs during antibody affinity maturation when immunoglobulin genes are hypermutated and only those B cells which express antibodies of improved antigen-binding specificity are expanded. Protein evolution might be simulated in cell culture, if transgene-specific hypermutation can be combined with the selection of cells carrying beneficial mutations. Here, we describe the optimization of a GFP transgene in the B cell line DT40 by hypermutation and iterative fluorescence activated cell sorting. Artificial evolution in DT40 offers unique advantages and may be easily adapted to other transgenes, if the selection for desirable mutations is feasible.
Figures


References
-
- Milstein C, Rada C. The maturation of the antibody response. In: Honjo T, Alt FW, editors. Immunoglobulin Genes. 2nd. London: Academic Press; 1995. pp. 57–81.
-
- Matsuura T, Yomo T. In vitro evolution of proteins. J. Biosci. Bioeng. 2006;101:449–456. - PubMed
-
- Dufner P, Jermutus L, Minter RR. Harnessing phage and ribosome display for antibody optimisation. Trends Biotechnol. 2006;24:523–529. - PubMed
-
- Dwyer MA, Looger LL, Hellinga HW. Computational design of a biologically active enzyme. Science. 2004;304:1967–1971. - PubMed
-
- Shimomura O. Structure of the chromophore of Aequorea green fluorescent protein. FEBS Lett. 1979;104:220–222.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

