Quantitative telomeric overhang determination using a double-strand specific nuclease
- PMID: 18073199
- PMCID: PMC2241892
- DOI: 10.1093/nar/gkm1063
Quantitative telomeric overhang determination using a double-strand specific nuclease
Abstract
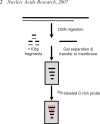
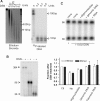
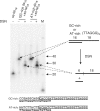
Telomeres terminate in 3' overhangs that function in end protection and the formation of t-loops. Determining the steps and factors involved in overhang processing is compromised by the inability to easily and accurately determine overhang size in the presence of many kilobases of double-stranded telomeric DNA. We here describe the use of a double-strand specific nuclease (DSN) that entirely digests double-stranded DNA including telomeres, leaving the overhangs intact so that they can be measured.
Figures

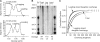
References
-
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. - PubMed
-
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. - PubMed
-
- Tahara H, Kusunoki M, Yamanaka Y, Matsumura S, Ide T. G-tail telomere HPA: simple measurement of human single-stranded telomeric overhangs. Nat. Methods. 2005;2:829–831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

