c-Met must translocate to the nucleus to initiate calcium signals
- PMID: 18073207
- PMCID: PMC2825875
- DOI: 10.1074/jbc.M706550200
c-Met must translocate to the nucleus to initiate calcium signals
Abstract
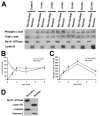
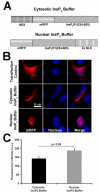
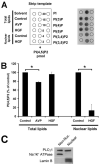
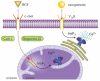
Hepatocyte growth factor (HGF) is important for cell proliferation, differentiation, and related activities. HGF acts through its receptor c-Met, which activates downstream signaling pathways. HGF binds to c-Met at the plasma membrane, where it is generally believed that c-Met signaling is initiated. Here we report that c-Met rapidly translocates to the nucleus upon stimulation with HGF. Ca(2+) signals that are induced by HGF result from phosphatidylinositol 4,5-bisphosphate hydrolysis and inositol 1,4,5-trisphosphate formation within the nucleus rather than within the cytoplasm. Translocation of c-Met to the nucleus depends upon the adaptor protein Gab1 and importin beta1, and formation of Ca(2+) signals in turn depends upon this translocation. HGF may exert its particular effects on cells because it bypasses signaling pathways in the cytoplasm to directly activate signaling pathways in the nucleus.
Figures

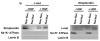

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

