Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes
- PMID: 18073417
- PMCID: PMC2219487
- DOI: 10.1534/genetics.107.080234
Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes
Abstract
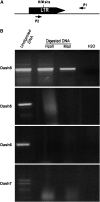
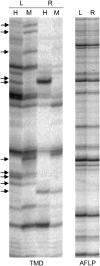
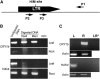
Transposable elements (TEs) represent approximately 45% of the human genome and 50-90% of some grass genomes. While most elements contain inactivating mutations, others are reversibly inactivated (silenced) by epigenetic mechanisms, including cytosine methylation. Previous studies have shown that retrotransposons can influence the expression of adjacent host genes. In this study, the methylation patterns of TEs and their flanking sequences in different tissues were undertaken using a novel technique called transposon methylation display (TMD). TMD was successfully applied on a highly copied (approximately 1000 copies), newly amplified LTR retrotransposon family in rice called Dasheng. We determined that the methylation status of a subset of LTRs varies in leaves vs. roots. In addition, we determined that tissue-specific LTR methylation correlated with tissue-specific expression of the flanking rice gene. Genes showing tissue-specific expression were in opposite orientation relative to the LTR. Antisense transcripts were detected in the tissue where the sense transcripts from that gene were not detected. Comparative analysis of Dasheng LTR methylation in the two subspecies, japonica vs. indica revealed LTR-mediated differences in subspecies gene expression. Subspecies-specific expression was due either to polymorphic Dasheng insertion sites between the two subspecies or to subspecies-specific methylation of LTRs at the same locus accounted for observed differences in the expression of adjacent genes.
Figures



Similar articles
-
Evolutionary history of Oryza sativa LTR retrotransposons: a preliminary survey of the rice genome sequences.BMC Genomics. 2004 Mar 2;5(1):18. doi: 10.1186/1471-2164-5-18. BMC Genomics. 2004. PMID: 15040813 Free PMC article.
-
LTR retrotransposons reveal recent extensive inter-subspecies nonreciprocal recombination in Asian cultivated rice.BMC Genomics. 2008 Nov 27;9:565. doi: 10.1186/1471-2164-9-565. BMC Genomics. 2008. PMID: 19038031 Free PMC article.
-
Evolutionary Epigenomics of Retrotransposon-Mediated Methylation Spreading in Rice.Mol Biol Evol. 2018 Feb 1;35(2):365-382. doi: 10.1093/molbev/msx284. Mol Biol Evol. 2018. PMID: 29126199 Free PMC article.
-
LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model.Cytogenet Genome Res. 2005;110(1-4):91-107. doi: 10.1159/000084941. Cytogenet Genome Res. 2005. PMID: 16093661 Review.
-
LTR-retrotransposons in plants: Engines of evolution.Gene. 2017 Aug 30;626:14-25. doi: 10.1016/j.gene.2017.04.051. Epub 2017 May 2. Gene. 2017. PMID: 28476688 Review.
Cited by
-
How important are transposons for plant evolution?Nat Rev Genet. 2013 Jan;14(1):49-61. doi: 10.1038/nrg3374. Nat Rev Genet. 2013. PMID: 23247435 Review.
-
Epigenetic interplay between mouse endogenous retroviruses and host genes.Genome Biol. 2012 Oct 3;13(10):R89. doi: 10.1186/gb-2012-13-10-r89. Genome Biol. 2012. PMID: 23034137 Free PMC article.
-
Transpositional reactivation of the Dart transposon family in rice lines derived from introgressive hybridization with Zizania latifolia.BMC Plant Biol. 2010 Aug 26;10:190. doi: 10.1186/1471-2229-10-190. BMC Plant Biol. 2010. PMID: 20796287 Free PMC article.
-
Polymorphism of a new Ty1-copia retrotransposon in durum wheat under salt and light stresses.Theor Appl Genet. 2010 Jul;121(2):311-22. doi: 10.1007/s00122-010-1311-z. Epub 2010 Mar 17. Theor Appl Genet. 2010. PMID: 20237753
-
Ty1-copia elements reveal diverse insertion sites linked to polymorphisms among flax (Linum usitatissimum L.) accessions.BMC Genomics. 2016 Dec 7;17(1):1002. doi: 10.1186/s12864-016-3337-3. BMC Genomics. 2016. PMID: 27927184 Free PMC article.
References
-
- Bennetzen, J. L., K. Schrick, P. S. Springer, W. E. Brown and P. Sanmiguel, 1994. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37: 565–576. - PubMed
-
- Chomczynski, P., 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 532. - PubMed
-
- Domansky, A. N., E. P. Kopantzev, E. V. Snezhkov, Y. B. Lebedev, C. Leib-Mosch et al., 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472: 191–195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

