The impact of genetic relationship information on genome-assisted breeding values
- PMID: 18073436
- PMCID: PMC2219482
- DOI: 10.1534/genetics.107.081190
The impact of genetic relationship information on genome-assisted breeding values
Abstract
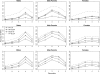
The success of genomic selection depends on the potential to predict genome-assisted breeding values (GEBVs) with high accuracy over several generations without additional phenotyping after estimating marker effects. Results from both simulations and practical applications have to be evaluated for this potential, which requires linkage disequilibrium (LD) between markers and QTL. This study shows that markers can capture genetic relationships among genotyped animals, thereby affecting accuracies of GEBVs. Strategies to validate the accuracy of GEBVs due to LD are given. Simulations were used to show that accuracies of GEBVs obtained by fixed regression-least squares (FR-LS), random regression-best linear unbiased prediction (RR-BLUP), and Bayes-B are nonzero even without LD. When LD was present, accuracies decrease rapidly in generations after estimation due to the decay of genetic relationships. However, there is a persistent accuracy due to LD, which can be estimated by modeling the decay of genetic relationships and the decay of LD. The impact of genetic relationships was greatest for RR-BLUP. The accuracy of GEBVs can result entirely from genetic relationships captured by markers, and to validate the potential of genomic selection, several generations have to be analyzed to estimate the accuracy due to LD. The method of choice was Bayes-B; FR-LS should be investigated further, whereas RR-BLUP cannot be recommended.
Figures


References
-
- Fernando, R. L., 1998. Genetic evaluation and selection using genotypic, phenotypic and pedigree information. Proceedings of the 6th World Congress on Genetics Applied to Livestock Production, Armidale, NSW, Australia, Vol. 26, pp. 329–336.
-
- Karlin, S., 1984. Theoretical aspects of genetic map functions in recombination processes, pp. 209–228 in Human Population Genetics: The Pittsburgh Symposium, edited by A. Chakravarti. Van Nostrand Reinhold, New York.
-
- Kutner, M. H., C. J. Nachtsheim, J. Neter and W. Li, 2005. Applied Linear Statistical Models, Ed. 5. McGraw-Hill, New York.
-
- Legarra, A., C. Robert-Granie, E. Manfredi and J. M. Elsen, 2007. Does genomic selection work in a mice population? Papers and Abstracts from the Workshop on QTL and Marker-Assisted Selection, edited by A. Legarra. March 22–23, 2007, Toulouse, France.
-
- Malécot, G., 1948. Les Mathématiques de l'Hérédité. Masson, Paris.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

