Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges
- PMID: 18074021
- PMCID: PMC2111050
- DOI: 10.1371/journal.pone.0001297
Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges
Abstract
Background: DNA helicases are ubiquitous enzymes that unwind DNA in an ATP-dependent and directionally specific manner. Unwinding of double-stranded DNA is essential for the processes of DNA repair, recombination, transcription, and DNA replication. Five human DNA helicases sharing sequence similarity with the E. coli RecQ helicase have been identified. Three of the human RecQ helicases are implicated in hereditary diseases (Bloom syndrome, Werner syndrome, and Rothmund-Thomson syndrome) which display clinical symptoms of premature aging and cancer. RECQ1 helicase is the most highly expressed of the human RecQ helicases; however, a genetic disease has yet not been linked to mutations in the RECQ1 gene, and the biological functions of human RECQ1 in cellular DNA metabolism are not known.
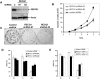
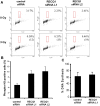
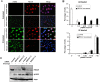
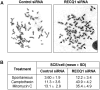
Methodology/principal findings: In this study, we report that RECQ1 becomes phosphorylated upon DNA damage and forms irradiation-induced nuclear foci that associate with chromatin in human cells. Depletion of RECQ1 renders human cells sensitive to DNA damage induced by ionizing radiation or the topoisomerase inhibitor camptothecin, and results in spontaneous gamma-H2AX foci and elevated sister chromatid exchanges, indicating aberrant repair of DNA breaks. Consistent with a role in homologous recombinational repair, endogenous RECQ1 is associated with the strand exchange protein Rad51 and the two proteins directly interact with high affinity.
Conclusion/significance: Collectively, these results provide the first evidence for a role of human RECQ1 in the response to DNA damage and chromosomal stability maintenance and point to the vital importance of RECQ1 in genome homeostasis.
Conflict of interest statement
Figures




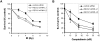

References
-
- Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. - PubMed
-
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. - PubMed
-
- Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S, Ozawa K, Eki T, Nogami M, Okumura K. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–4573. - PMC - PubMed
-
- Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

