Prion strain discrimination in cell culture: the cell panel assay
- PMID: 18077360
- PMCID: PMC2409240
- DOI: 10.1073/pnas.0710054104
Prion strain discrimination in cell culture: the cell panel assay
Abstract
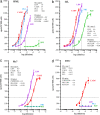
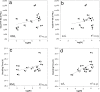
Prions are thought to consist mainly or entirely of misfolded PrP, a constitutively expressed host protein. Prions associated with the same PrP sequence may occur in the form of different strains; the strain phenotype is believed to be encoded by the conformation of the PrP. Some cell lines can be persistently infected by prions and, interestingly, show preference for certain strains. We report that a cloned murine neuroblastoma cell population, N2a-PK1, is highly heterogeneous in regard to its susceptibility to RML and 22L prions. Remarkably, sibling subclones may show very different relative susceptibilities to the two strains, indicating that the responses can vary independently. We have assembled four cell lines, N2a-PK1, N2a-R33, LD9 and CAD5, which show widely different responses to prion strains RML, 22L, 301C, and Me7, into a panel that allows their discrimination in vitro within 2 weeks, using the standard scrapie cell assay (SSCA).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Unraveling prion strains with cell biology and organic chemistry.Proc Natl Acad Sci U S A. 2008 Jan 8;105(1):11-2. doi: 10.1073/pnas.0710824105. Epub 2008 Jan 2. Proc Natl Acad Sci U S A. 2008. PMID: 18172195 Free PMC article. No abstract available.
References
-
- Prusiner SB. Annu Rev Microbiol. 1989;43:345–374. - PubMed
-
- Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A. Biochemistry. 2002;41:12868–12875. - PubMed
-
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Nat Med. 1998;4:1157–1165. - PubMed
-
- Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR. Biochemistry. 2006;45:15710–15717. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

