A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway
- PMID: 18077367
- PMCID: PMC2154428
- DOI: 10.1073/pnas.0707999105
A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway
Abstract
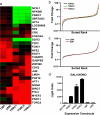
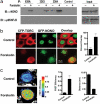
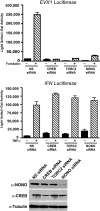
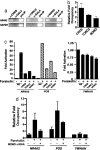
Signal transduction pathways often use a transcriptional component to mediate adaptive cellular responses. Coactivator proteins function prominently in these pathways as the conduit to the basic transcriptional machinery. Here we present a high-throughput cell-based screening strategy, termed the "coactivator trap," to study the functional interactions of coactivators with transcription factors. We applied this strategy to the cAMP signaling pathway, which utilizes two families of coactivators, the cAMP response element binding protein (CREB) binding protein (CBP)/p300 family and the recently identified transducers of regulated CREB activity family (TORCs1-3). In addition to identifying numerous known interactions of these coactivators, this analysis identified NONO (p54(nrb)) as a TORC-interacting protein. RNA interference experiments demonstrate that NONO is necessary for cAMP-dependent activation of CREB target genes in vivo. Furthermore, TORC2 and NONO complex on cAMP-responsive promoters, and NONO acts as a bridge between the CREB/TORC complex and RNA polymerase II. These data demonstrate the utility of the coactivator trap by identification of a component of cAMP-mediated transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rosenfeld MG, Lunyak VV, Glass CK. Genes Dev. 2006;20:1405–1428. - PubMed
-
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Nature. 1993;365:855–859. - PubMed
-
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. Mol Cell. 2003;12:413–423. - PubMed
-
- Gonzalez GA, Montminy MR. Cell. 1989;59:675–680. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

