Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage
- PMID: 18077395
- PMCID: PMC2410075
- DOI: 10.1073/pnas.0710061104
Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage
Abstract
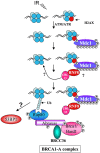
The Brca1 A complex contains Brca1/Bard1, Abraxas, Rap80, and Brcc36; however, with the exception of the Brca1-Abraxas interaction, how the A complex is assembled is not known. The A complex is localized to sites of DNA damage through the UIM domains of RAP80, which bind K63-linked polyubiquitin chains. In this study, we identified an FHA domain RING finger E3 ubiquitin ligase, RNF8, and an E2-conjugating enzyme known to form K63-polyubiquitin chains, Ubc13, each of which is required to recruit the Brca1 A complex to sites of DNA damage. Rnf8 localizes to sites of DNA damage through an FHA-domain-containing region. We found that Rap80 contains an Abraxas interaction domain [AIR (Abraxas-interacting region)], required for association of Rap80 with Abraxas, Brca1, and Brcc36. Abraxas and Brcc36 associate through coiled-coil domains on each protein. These data suggest a model through which Ubc13 and Rnf8 are recruited to sites of DNA damage through DNA-damage-induced phosphorylation of a chromatin-associated protein and generate polyubiquitin chains that then recruit Rap80 and the entire Brca1 A complex to DNA-damage foci. This sequential E3 ubiquitin ligase recruitment constitutes a ubiquitin ligase cascade required for DNA repair and checkpoint signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Launching a ubiquitination cascade at DNA breaks.Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20645-6. doi: 10.1073/pnas.0710916105. Epub 2007 Dec 19. Proc Natl Acad Sci U S A. 2007. PMID: 18093907 Free PMC article. No abstract available.
References
-
- Venkitaraman AR. Cell. 2002;108:171–182. - PubMed
-
- Narod SA, Foulkes WD. Nat Rev Cancer. 2004;4:665–676. - PubMed
-
- Baer R, Ludwig T. Curr Opin Genet Dev. 2002;12:86–91. - PubMed
-
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Nat Genet. 1996;14:430–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

