Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast
- PMID: 18077551
- PMCID: PMC2230582
- DOI: 10.1091/mbc.e07-09-0968
Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast
Abstract
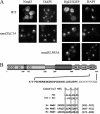
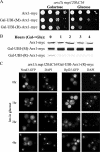
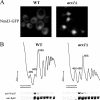
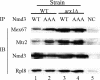
We previously showed that nuclear export of the large (60S) ribosomal subunit relies on Nmd3 in a Crm1-dependent manner. Recently the general mRNA export factor, the Mtr2/Mex67 heterodimer, was shown to act as an export receptor in parallel with Crm1. These observations raise the possibility that nuclear export of the 60S subunit in Saccharomyces cerevisiae requires multiple export receptors. Here, we show that the previously characterized 60S subunit biogenesis factor, Arx1, also acts as an export receptor for the 60S subunit. We found that deletion of ARX1 was synthetic lethal with nmd3 and mtr2 mutants and was synthetic sick with several nucleoporin mutants. Deletion of ARX1 led to accumulation of pre-60S particles in the nucleus that were enriched for Nmd3, Crm1, Mex67, and Mtr2, suggesting that in the absence of Arx1, 60S export is impaired even though the subunit is loaded with export receptors. Finally, Arx1 interacted with several nucleoporins in yeast two-hybrid as well as in vitro assays. These results show that Arx1 can directly bridge the interaction between the pre-60S particle and the NPC and thus is a third export receptor for the 60S subunit in yeast.
Figures


Similar articles
-
Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae.Mol Cell Biol. 2006 May;26(10):3718-27. doi: 10.1128/MCB.26.10.3718-3727.2006. Mol Cell Biol. 2006. PMID: 16648468 Free PMC article.
-
Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit.Mol Cell. 2007 Sep 7;27(5):767-79. doi: 10.1016/j.molcel.2007.06.034. Mol Cell. 2007. PMID: 17803941
-
Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex.J Biol Chem. 2007 May 11;282(19):14028-37. doi: 10.1074/jbc.M700256200. Epub 2007 Mar 8. J Biol Chem. 2007. PMID: 17347149
-
Nuclear export of ribosomal subunits.Trends Biochem Sci. 2002 Nov;27(11):580-5. doi: 10.1016/s0968-0004(02)02208-9. Trends Biochem Sci. 2002. PMID: 12417134 Review.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
-
The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits.EMBO Rep. 2011 Sep 30;12(10):1024-31. doi: 10.1038/embor.2011.155. EMBO Rep. 2011. PMID: 21852791 Free PMC article.
-
Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1.Nat Struct Mol Biol. 2017 Mar;24(3):214-220. doi: 10.1038/nsmb.3364. Epub 2017 Jan 23. Nat Struct Mol Biol. 2017. PMID: 28112732 Free PMC article.
-
The T-cell leukemia related rpl10-R98S mutant traps the 60S export adapter Nmd3 in the ribosomal P site in yeast.PLoS Genet. 2017 Jul 17;13(7):e1006894. doi: 10.1371/journal.pgen.1006894. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28715419 Free PMC article.
-
Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines.Curr Opin Cell Biol. 2009 Feb;21(1):109-18. doi: 10.1016/j.ceb.2009.01.003. Epub 2009 Jan 21. Curr Opin Cell Biol. 2009. PMID: 19167202 Free PMC article. Review.
-
Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes.PLoS Genet. 2012;8(8):e1002915. doi: 10.1371/journal.pgen.1002915. Epub 2012 Aug 30. PLoS Genet. 2012. PMID: 22956913 Free PMC article.
References
-
- Allen N. P., Huang L., Burlingame A., Rexach M. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 2001;276:29268–29274. - PubMed
-
- Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell Proteom. 2002;1:930–946. - PubMed
-
- Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

