Genome organization and reverse genetic analysis of a type I feline coronavirus
- PMID: 18077720
- PMCID: PMC2258703
- DOI: 10.1128/JVI.02339-07
Genome organization and reverse genetic analysis of a type I feline coronavirus
Abstract
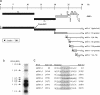
In this study we report the complete sequence and genome organization of the serotype I feline coronavirus (FCoV) strain Black. Furthermore, a reverse genetic system was established for this FCoV strain by cloning a full-length cDNA copy into vaccinia virus. This clone served as basis for the generation of recombinant FCoV (recFCoV) that was shown to bear the same features in vitro as the parental FCoV. Using this system, accessory 3abc genes in the FCoV genome were replaced by green fluorescent protein (recFCoV-GFP) and Renilla luciferase genes (recFCoV-RL). In addition, we showed that feline CD14(+) blood monocytes and dendritic cells can be easily detected after infection with recFCoV-GFP. Thus, our established reverse genetic system provides a suitable tool to study the molecular biology of serotype I FCoV.
Figures


References
-
- Addie, D. D., and O. Jarrett. 2001. Use of a reverse-transcriptase polymerase chain reaction for monitoring the shedding of feline coronavirus by healthy cats. Vet. Rec. 148649-653. - PubMed
-
- Addie, D. D., I. A. Schaap, L. Nicolson, and O. Jarrett. 2003. Persistence and transmission of natural type I feline coronavirus infection. J. Gen. Virol. 842735-2744. - PubMed
-
- Black, J. W. 1980. Recovery and in vitro cultivation of a coronavirus from laboratory-induced cases of feline infectious peritonitis (FIP). Vet. Med. Small Anim. Clin. 75811-814. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

