Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae
- PMID: 18079178
- PMCID: PMC2113032
- DOI: 10.1101/gad.449407
Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae
Abstract
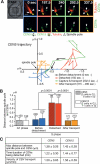
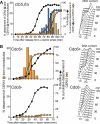
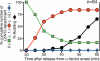
In the budding yeast Saccharomyces cerevisiae, microtubule-organizing centers called spindle pole bodies (SPBs) are embedded in the nuclear envelope, which remains intact throughout the cell cycle (closed mitosis). Kinetochores are tethered to SPBs by microtubules during most of the cell cycle, including G1 and M phases; however, it has been a topic of debate whether microtubule interaction is constantly maintained or transiently disrupted during chromosome duplication. Here, we show that centromeres are detached from microtubules for 1-2 min and displaced away from a spindle pole in early S phase. These detachment and displacement events are caused by centromere DNA replication, which results in disassembly of kinetochores. Soon afterward, kinetochores are reassembled, leading to their recapture by microtubules. We also show how kinetochores are subsequently transported poleward by microtubules. Our study gives new insights into kinetochore-microtubule interaction and kinetochore duplication during S phase in a closed mitosis.
Figures





References
-
- Adams I.R., Kilmartin J.V., Kilmartin J.V. Spindle pole body duplication: A model for centrosome duplication? Trends Cell Biol. 2000;10:329–335. - PubMed
-
- Amberg D.C., Burke D.J., Strathern J.N., Burke D.J., Strathern J.N., Strathern J.N. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005.
-
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Sassoon I., Hyman A.A., Murray A.W., Hyman A.A., Murray A.W., Murray A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. - PMC - PubMed
-
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
