TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis
- PMID: 18079962
- PMCID: PMC2129232
- DOI: 10.1172/JCI29900
TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis
Abstract
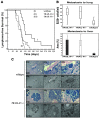
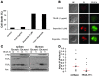
Preclinical data support the potential of the death-signaling receptors for TRAIL as targets for cancer therapy. However, it is unclear whether these death-signaling receptors suppress the emergence and growth of malignant tumors in vivo. Herein we show that TNF-related apoptosis-inducing ligand receptor (TRAIL-R), the only proapoptotic death-signaling receptor for TRAIL in the mouse, suppresses inflammation and tumorigenesis. Loss of a single TRAIL-R allele on the lymphoma-prone Emu-myc genetic background significantly reduced median lymphoma-free survival. TRAIL-R-deficient lymphomas developed with equal frequency irrespective of mono- or biallelic loss of TRAIL-R, had increased metastatic potential, and showed apoptotic defects relative to WT littermates. In addition, TRAIL-R-/- mice showed decreased long-term survival following a sublethal dose of ionizing radiation. Histological evaluation of moribund irradiated TRAIL-R-/- animals showed hallmarks of bronchopneumonia as well as tumor formation with increased NF-kappaB p65 expression. TRAIL-R also suppressed diethylnitrosamine-induced (DEN-induced) hepatocarcinogenesis, as an increased number of large tumors with apoptotic defects developed in the livers of DEN-treated TRAIL-R-/- mice. Thus TRAIL-R may function as an inflammation and tumor suppressor in multiple tissues in vivo.
Figures






Comment in
-
TRAIL-R deficiency in mice enhances lymph node metastasis without affecting primary tumor development.J Clin Invest. 2008 Jan;118(1):100-10. doi: 10.1172/JCI33061. J Clin Invest. 2008. PMID: 18079967 Free PMC article.
References
-
- Boldin M.P., et al. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 1995;270:7795–7798. - PubMed
-
- Chinnaiyan A.M., O’Rourke K., Tewari M., Dixit V.M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. - PubMed
-
- Bodmer J.L., et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2000;2:241–243. - PubMed
-
- Kischkel F.C., et al. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. - PubMed
-
- Sprick M.R., et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

