Understanding the dynamic behavior of genetic regulatory networks by functional decomposition
- PMID: 18079985
- PMCID: PMC2134916
- DOI: 10.2174/138920206778948718
Understanding the dynamic behavior of genetic regulatory networks by functional decomposition
Abstract
A number of mechanistic and predictive genetic regulatory networks (GRNs) comprising dozens of genes have already been characterized at the level of cis-regulatory interactions. Reconstructions of networks of 100's to 1000's of genes and their interactions are currently underway. Understanding the organizational and functional principles underlying these networks is probably the single greatest challenge facing genomics today. We review the current approaches to deciphering large-scale GRNs and discuss some of their limitations. We then propose a bottom-up approach in which large-scale GRNs are first organized in terms of functionally distinct GRN building blocks of one or a few genes. Biological processes may then be viewed as the outcome of functional interactions among these simple, well-characterized functional building blocks. We describe several putative GRN functional building blocks and show that they can be located within GRNs on the basis of their interaction topology and additional, simple and experimentally testable constraints.
Keywords: Genetic regulatory networks; systems biology; transcriptional regulation; visualization.
Figures
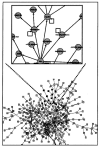




References
-
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. - PubMed
-
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane–Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. - PMC - PubMed
-
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, Tissenbaum HA, Mango SE, Walhout AJ. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–205. - PubMed
-
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Elsevier; 2006.
-
- Ihmels J, Bergmann S, Barkai N. Defining transcription modules using large-scale gene expression data. Bioinformatics. 2004;20:1993–2003. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
