Drosophila telomeres: an exception providing new insights
- PMID: 18081009
- PMCID: PMC2804870
- DOI: 10.1002/bies.20688
Drosophila telomeres: an exception providing new insights
Abstract
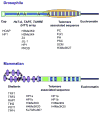
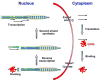
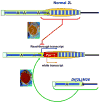
Drosophila telomeres comprise DNA sequences that differ dramatically from those of other eukaryotes. Telomere functions, however, are similar to those found in telomerase-based telomeres, even though the underlying mechanisms may differ. Drosophila telomeres use arrays of retrotransposons to maintain chromosome length, while nearly all other eukaryotes rely on telomerase-generated short repeats. Regardless of the DNA sequence, several end-binding proteins are evolutionarily conserved. Away from the end, the Drosophila telomeric and subtelomeric DNA sequences are complexed with unique combinations of proteins that also modulate chromatin structure elsewhere in the genome. Maintaining and regulating the transcriptional activity of the telomeric retrotransposons in Drosophila requires specific chromatin structures and, while telomeric silencing spreads from the terminal repeats in yeast, the source of telomeric silencing in Drosophila is the subterminal arrays. However, the subterminal arrays in both species may be involved in telomere-telomere associations and/or communication.
(c) 2007 Wiley Periodicals, Inc.
Figures
Similar articles
-
Drosophila telomeres: the non-telomerase alternative.Chromosome Res. 2005;13(5):431-41. doi: 10.1007/s10577-005-0992-7. Chromosome Res. 2005. PMID: 16132809 Review.
-
Structure of telomeric chromatin in Drosophila.Biochemistry (Mosc). 2007 Jun;72(6):618-30. doi: 10.1134/s0006297907060041. Biochemistry (Mosc). 2007. PMID: 17630906 Review.
-
Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres.Annu Rev Genet. 2003;37:485-511. doi: 10.1146/annurev.genet.38.072902.093115. Annu Rev Genet. 2003. PMID: 14616071 Review.
-
Drosophila telomeres: A variation on the telomerase theme.Fly (Austin). 2008 May-Jun;2(3):101-10. doi: 10.4161/fly.6393. Epub 2008 May 4. Fly (Austin). 2008. PMID: 18820466 Review.
-
Drosophila telomeric retrotransposons derived from an ancestral element that was recruited to replace telomerase.Genome Res. 2007 Dec;17(12):1909-18. doi: 10.1101/gr.6365107. Epub 2007 Nov 7. Genome Res. 2007. PMID: 17989257 Free PMC article.
Cited by
-
Rapid evolution at the Drosophila telomere: transposable element dynamics at an intrinsically unstable locus.Genetics. 2021 Feb 9;217(2):iyaa027. doi: 10.1093/genetics/iyaa027. Genetics. 2021. PMID: 33724410 Free PMC article.
-
Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity.Front Genet. 2015 Apr 14;6:143. doi: 10.3389/fgene.2015.00143. eCollection 2015. Front Genet. 2015. PMID: 25926849 Free PMC article. Review.
-
Curiously composite structures of a retrotransposon and a complex repeat associated with chromosome ends of Rhynchosciara americana (Diptera: Sciaridae).Chromosome Res. 2010 Jul;18(5):587-98. doi: 10.1007/s10577-010-9143-x. Epub 2010 Jul 7. Chromosome Res. 2010. PMID: 20607598
-
Unusually short tandem repeats appear to reach chromosome ends of Rhynchosciara americana (Diptera: Sciaridae).Chromosoma. 2010 Dec;119(6):613-23. doi: 10.1007/s00412-010-0282-9. Epub 2010 Jul 8. Chromosoma. 2010. PMID: 20614221
-
CriTER-A: A Novel Temperature-Dependent Noncoding RNA Switch in the Telomeric Transcriptome of Chironomus riparius.Int J Mol Sci. 2021 Sep 24;22(19):10310. doi: 10.3390/ijms221910310. Int J Mol Sci. 2021. PMID: 34638651 Free PMC article.
References
-
- Mason JM, Biessmann H. The unusual telomeres of Drosophila. Trends Genet. 1995;11:58–62. - PubMed
-
- Louis EJ, Vershinin AV. Chromosome ends: different sequences may provide conserved functions. Bioessays. 2005;27:685–697. - PubMed
-
- Muller HJ. The remaking of chromosomes. The Collecting Net. 1938;8:182–195.
-
- Watson JD. Origin of concatameric T7. DNA Nature New Biol. 1972;239:197–201. - PubMed
-
- Olovnikov AM. A theory of marginotomy. J Theor Biol. 1973;41:181–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

