AGEMAP: a gene expression database for aging in mice
- PMID: 18081424
- PMCID: PMC2098796
- DOI: 10.1371/journal.pgen.0030201
AGEMAP: a gene expression database for aging in mice
Abstract
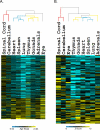
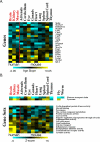
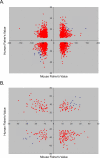
We present the AGEMAP (Atlas of Gene Expression in Mouse Aging Project) gene expression database, which is a resource that catalogs changes in gene expression as a function of age in mice. The AGEMAP database includes expression changes for 8,932 genes in 16 tissues as a function of age. We found great heterogeneity in the amount of transcriptional changes with age in different tissues. Some tissues displayed large transcriptional differences in old mice, suggesting that these tissues may contribute strongly to organismal decline. Other tissues showed few or no changes in expression with age, indicating strong levels of homeostasis throughout life. Based on the pattern of age-related transcriptional changes, we found that tissues could be classified into one of three aging processes: (1) a pattern common to neural tissues, (2) a pattern for vascular tissues, and (3) a pattern for steroid-responsive tissues. We observed that different tissues age in a coordinated fashion in individual mice, such that certain mice exhibit rapid aging, whereas others exhibit slow aging for multiple tissues. Finally, we compared the transcriptional profiles for aging in mice to those from humans, flies, and worms. We found that genes involved in the electron transport chain show common age regulation in all four species, indicating that these genes may be exceptionally good markers of aging. However, we saw no overall correlation of age regulation between mice and humans, suggesting that aging processes in mice and humans may be fundamentally different.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. - PubMed
-
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. - PubMed
-
- Edwards MG, Sarkar D, Klopp R, Morrow JD, Weindruch R, et al. Impairment of the transcriptional responses to oxidative stress in the heart of aged C57BL/6 mice. Ann N Y Acad Sci. 2004;1019:85–95. - PubMed
-
- Dhahbi JM, Tsuchiya T, Kim HJ, Mote PL, Spindler SR. Gene expression and physiologic responses of the heart to the initiation and withdrawal of caloric restriction. J Gerontol A Biol Sci Med Sci. 2006;61:218–231. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

